Sample Size Calculation under Non-Proportional Hazards.
package: nphPower
Package: nphPower provides functions to perform combination test including maximum weighted logrank test (MWLR) and projection test, to calculate sample size with MWLR in a simulation-free approach allowing for staggered entry, drop-out etc, to visualize the design parameters and to simulate survival data with flexible design input.
Installation
You can install the released version of nphPower from CRAN with:
install.packages("nphPower")
library("nphPower")
And the development version from GitHub with:
# install.packages("devtools")
#devtools::install_github("hcheng99/nphPower")
Example 1 - perform maximum weighted logrank test
This is a basic example which shows you how to perform maximum weighted logrank test.
Load the lung data and only keep columns for analysis.
lung <- nphPower::lung
tmpd <- with(lung, data.frame(time = SurvTime, stat = 1-censor, grp = Treatment))
Generate the weight functions for maxcombo test
wmax <- gen.wgt(method = "Maxcombo")
Perform the test using pooled Kaplan-Meier estimate of CDF as base function and visualize the weight functions
t1 <- MaxLRtest(tmpd, Wlist = wmax, base = c("KM"),
alternative = c("two.sided"))
plot(t1)
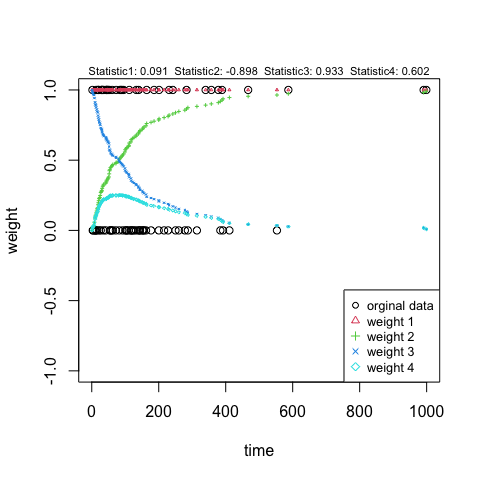 ## Example 2 - sample size under proportional hazard The design setting is: 5 years’ entry time and 5 years’ follow-up time; Median survival for control group is 10 years. The hazard ratio comparing treatment and control is 0.5.
## Example 2 - sample size under proportional hazard The design setting is: 5 years’ entry time and 5 years’ follow-up time; Median survival for control group is 10 years. The hazard ratio comparing treatment and control is 0.5.
t_enrl <- 5; t_fup <- 5 ; lmd0 <- -log(0.2)/10 ; HR <- 0.5
eg1 <- pwr2n.LR(method = "schoenfeld", lambda0 = lmd0,
lambda1 = lmd0*HR, entry = t_enrl, fup = t_fup)
#> ------------------------------------------
#> -----Summary of the Input Parameters-----
#> ------------------------------------------
#> __Parameter__ __Value__
#> Method schoenfeld
#> Lambda1/Lambda0/HR 0.08/0.161/0.5
#> Entry Time 5
#> Follow-up Time 5
#> Allocation Ratio 1
#> Type I Error 0.05
#> Type II Error 0.1
#> Alternative two.sided
#> Drop-out Parameter Not Provided
#> ------------------------------------------
#> -----Summary of the Output Parameters-----
#> ------------------------------------------
#> __Parameter__ __Value__
#> Number of Events 87.479
#> Number of Total Sampe Size 153.173
#> Overall Event Rate 0.571
Example 3 - sample size under nonproportional hazard
Design setting: patients are enrolled within 12 months and the last enrolled patient has at least 18 months’ follow-up. The medial survival time for control group is 12 months. The treatment has delayed effects. The hazard ratio is 0.75 after 6 months. Maxcombo test is used.
t_enrl <- 12; t_fup <- 18; lmd0 <- log(2)/12
f_hr_delay <- function(x){(x<=6)+(x>6)*0.75}
f_haz0 <- function(x){lmd0*x^0}
snph1 <- pwr2n.NPH(entry = t_enrl, fup = t_fup, Wlist = wmax,
k = 50, ratio = 2, CtrlHaz = f_haz0, hazR = f_hr_delay)
#> -----Summary of the Input Parameters-----
#> parameter value
#> Method MaxLR
#> Entry Time 12
#> Follow-up Time 18
#> Allocation Ratio 2
#> Type I Error 0.05
#> Type II Error 0.1
#> Alternative two.sided
#> Number of Weights 4
#> -----Summary of the Output Parameters-----
#> parameter value
#> Number of Events 1198.779
#> Number of Total Sampe Size 1720.967
#> Asymptotic Power 0.900
#> Overall Event Rate 0.697
Example 4 - trial data simulation
A time-to-event data set with settings in example 3 is simulated.
N <- round(snph1$totalN, digits = 0)
set.seed(12345)
simu1 <- simu.trial(type = "time", trial_param = c(N,t_enrl,
t_fup), bsl_dist = "weibull", bsl_param = c(1,lmd0),
HR_fun = f_hr_delay, ratio = 1)
#> Notes: Drop-outs are not considered in the simulation.
#> -------- Summary of the Simulation --------
#> parameter value
#> 1 Trial Type: time
#> 2 Entry Time: 12
#> 3 Maximum Study Duration: 30
#> 4 Number of Subjects: 1721
#> 5 Number of Events: 1259
More functions can be found in the package.