Description
Precision Agriculture Data Analysis.
Description
Precision agriculture spatial data depuration and homogeneous zones (management zone) delineation. The package includes functions that performs protocols for data cleaning management zone delineation and zone comparison; protocols are described in Paccioretti et al., (2020) <doi:10.1016/j.compag.2020.105556>.
README.md
paar 
The goal of paar is to provide useful functions for precision agriculture spatial data depuration.
Installation
You can install the released version of paar from CRAN with (not-yet):
# install.packages("paar")
You can install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("PPaccioretti/paar")
Example
The package has a complete protocol for automating error removal. Default values of all functions are optimized for precision agricultural data.
library(paar)
library(sf)
#> Warning: package 'sf' was built under R version 4.3.3
data("barley", package = 'paar')
barley data contains barley grain yield which were obtained using calibrated commercial yield monitors, mounted on combines equipped with DGPS.
#Convert barley data to an spatial object
barley_sf <- st_as_sf(barley,
coords = c("X", "Y"),
crs = 32720)
barley_dep <-
depurate(barley_sf,
"Yield")
#> Concave hull algorithm is computed with
#> concavity = 2 and length_threshold = 0
# Summary of depurated data
summary(barley_dep)
#> normal point border spatial outlier MP spatial outlier LM
#> 5673 (77%) 964 (13%) 343 (4.6%) 309 (4.2%)
#> global min outlier
#> 99 (1.3%) 6 (0.081%)
Spatial yield values before and after depuration process can be plotted
plot(barley_sf["Yield"], main = "Before depuration")
plot(barley_dep$depurated_data["Yield"], main = "After depuration")
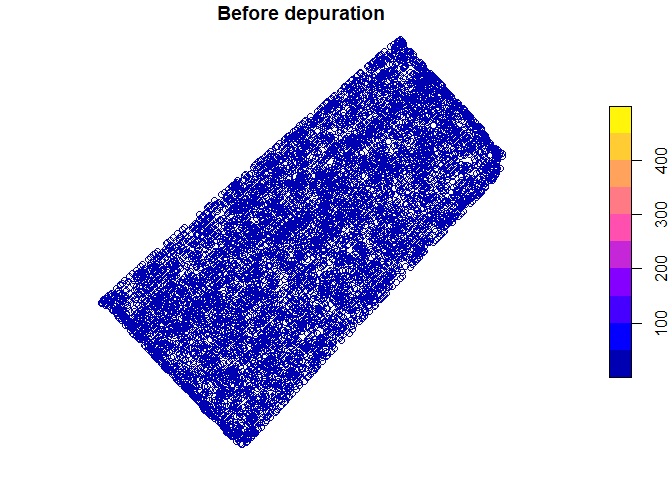
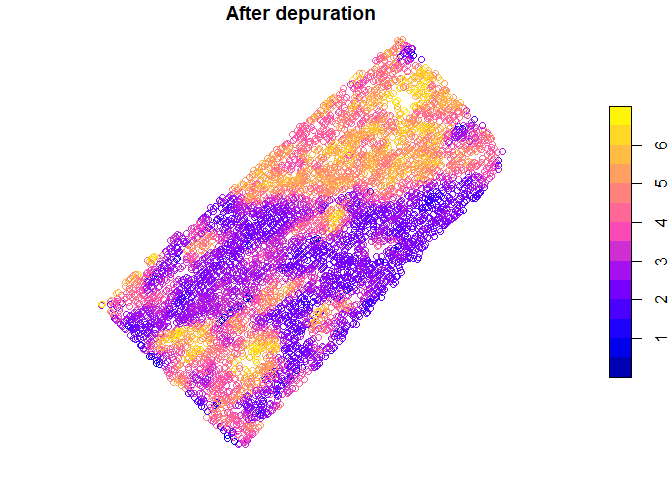
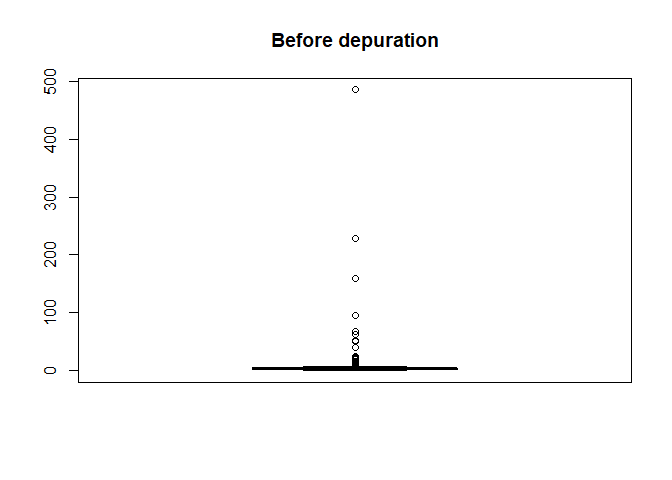
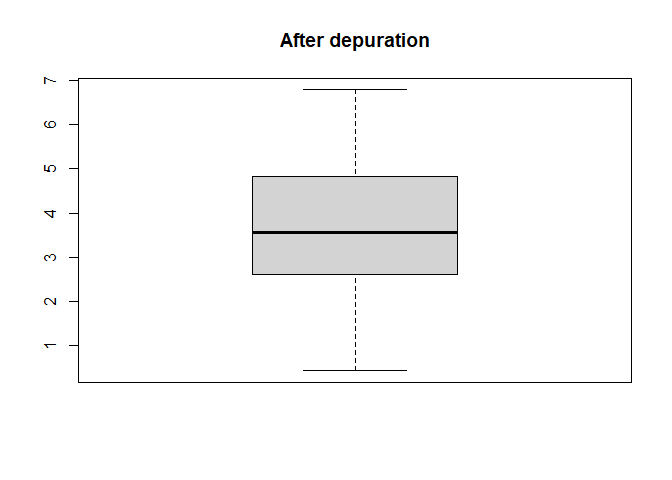
Also distribution of yield values can be plotted
boxplot(barley_sf[["Yield"]], main = "Before depuration")
boxplot(barley_dep$depurated_data[["Yield"]], main = "After depuration")
References
- Vega, A., Córdoba, M., Castro-Franco, M. et al. (2019). Protocol for automating error removal from yield maps. Precision Agric 20, 1030–1044 https://doi.org/10.1007/s11119-018-09632-8
- Paccioretti, P., Córdoba, M., & Balzarini, M. (2020). FastMapping: Software to create field maps and identify management zones in precision agriculture. Computers and Electronics in Agriculture, 175, 105556 https://doi.org/10.1016/j.compag.2020.105556.
