Simulating Diversification Dynamics.
paleobuddy
paleobuddy is an R package to simulate species diversification, fossil records, and phylogenetic trees. While the literature on species birth-death simulators is extensive, including important software like paleotree and APE, we concluded there were interesting gaps to be filled regarding possible diversification scenarios. Differently from most simulators in the field, we strove for flexibility over focus, implementing a large array of regimens for users to experiment with and combine, and structuring the package on a general framework to allow for straightforward expansion of available scenarios. In this way, paleobuddy can be used as a complement to other simulators or, in the case of scenarios implemented only here, can allow for robust and easy simulations for novel scenarios.
Installation
You can install the released version of paleobuddy from CRAN with:
install.packages("paleobuddy")
And the development version from GitHub with:
library(devtools)
devtools::install_github("brpetrucci/paleobuddy")
Example
We run a simple birth-death simulation as follows
set.seed(1)
n0 <- 1 # initial number of species
lambda <- 0.1 # speciation rate
mu <- 0.05 # extinction rate
tMax <- 30 # maximum simulation time
# run simulation
sim <- bd.sim(n0, lambda, mu, tMax)
We can then generate fossil records, and visualize the results
set.seed(1)
rho <- 1 # sampling rate
bins <- seq(tMax, 0, -1) # something to simulate geologic intervals
# get a data frame with fossil occurrence times
fossils <- sample.clade(sim = sim, rho = rho, tMax = tMax, bins = bins)
# visualize simulation and fossil occurrences
draw.sim(sim, fossils = fossils)
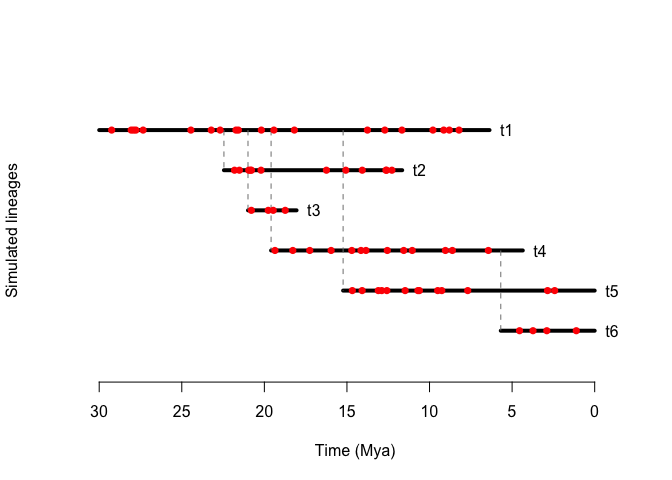
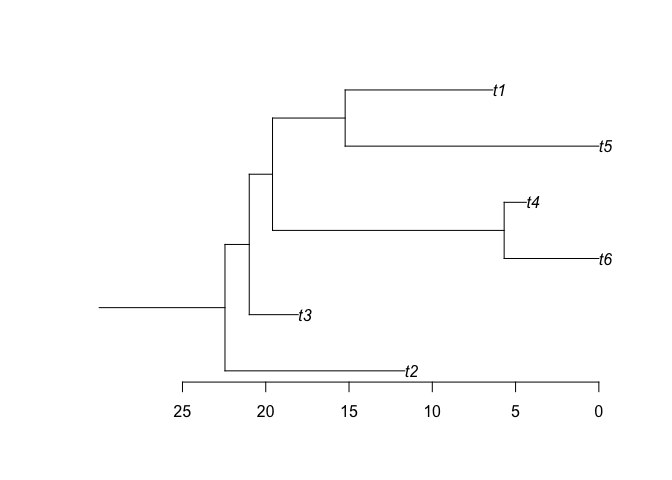
And generate phylogenies as well
phy <- make.phylo(sim) # make a phylogenetic tree with the simulated group
ape::plot.phylo(phy, root.edge = TRUE) # plot it with a stem (requires APE)
ape::axisPhylo() # add axis
Important functions
bd.sim is the birth-death simulation function, allowing for multiple arguments to build a large number of possible scenarios. One can supply constant or time-dependent speciation rate lambda and extinction rate mu. On top of the base rates, we allow for a shape parameter for each, if one chooses to interpret lambda and mu as scales of a Weibull distribution for age-dependent diversification. We take the novel step allowing for time-dependent scale and shape as well. One can also supply an env parameter to make rates dependent on a time-series, such as temperature. These can all be combined as the user wishes, creating a myriad of possible scenarios.
sample.clade generates fossil records, returning an organized data frame with occurrence times - or occurrence time ranges, provided the user supplies the respective interval vector. It allows for a sampling rate rho that can be as flexible as lambda and mu above, with the exception of a shape parameter, since we omitted that option given the absence of the use of Weibull distributions to model age-dependent fossil sampling in the literature. Instead, we allow for the user to supply a function they wish to use as age-dependent sampling, adFun, such as the PERT distribution used in PyRate.
bd.sim.traits and sample.clade.traits work similarly to bd.sim and sample.clade, but on the context of state-dependent diversification, in particular using the MuSSE model.
make.phylo closes the trio of most important functions of the package, taking a paleobuddy simulation and returning a phylo object from the APE package (see above).
draw.sim allows for easy visualization of birth-death simulation objects, drawing species’ durations and kinship, besides allowing for the addition of fossil occurrences as well.
Besides its main simulating and visualization functions, paleobuddy also supplies the user with a few interesting statistical tools, such as rexp.var, a generalization of the rexp function in base R that allows for time-varying exponential rates and a shape parameter, in which case it generalizes the rweibull function.
Data
Given the possibility of functions in paleobuddy to use environmentally-dependent rates, we have included with the package data frames containing environmental data, namely temperature (temp) and co2 (co2). These have been modified from data on RPANDA (RPANDA: Morlon H. et al (2016) RPANDA: an R package for macroevolutionary analyses on phylogenetic trees. Methods in Ecology and Evolution 7: 589-597). To see more about the origin of the data, see ?data, where data is the data frame’s name.
Authors
paleobuddy was idealized by Bruno do Rosario Petrucci and Tiago Bosisio Quental. The birth-death, statistical, and part of the sampling functions were written by Bruno. The phylogeny and most of the sampling functions were written by Matheus Januário.