Import and Export 'Familias' Files.
pedFamilias
The purpose of pedFamilias is to facilitate data exchanging between the pedsuite packages and the Familias software for forensic kinship analysis. The main functions are readFam() and writeFam() for reading and writing .fam files, which are used by Familias.
The readFam() function also supports files written with the DVI module of Familias, and also pure database files without pedigree information.
These functions were originally part of forrel, but were split out to streamline maintenance and provide a more lightweight import for other packages otherwise independent of forrel.
Installation
Install pedFamilias from CRAN as follows:
install.packages("pedFamilias")
Alternatively, install the development version from GitHub:
# install.packages("remotes")
remotes::install_github("magnusdv/pedFamilias")
Example
library(pedFamilias)
For a simple illustration of pedFamilias we read the example file paternity.fam shipped with the package:
fam = system.file("extdata", "paternity.fam", package = "pedFamilias")
peds = readFam(fam)
#> Familias version: 3.3.1
#> Read DVI: No
#>
#> Number of individuals (excluding 'extras'): 3
#> Individual 'AF': Genotypes for 10 markers read
#> Individual 'MO': Genotypes for 0 markers read
#> Individual 'CH': Genotypes for 10 markers read
#>
#> Number of pedigrees: 2
#> Pedigree 'H1' (0 extra females, 0 extra males)
#> Pedigree 'H2' (0 extra females, 0 extra males)
#>
#> Database: unknown
#> Number of loci: 10
#> D3S1358: 12 alleles, mut model (M/F) = step-ext, rate = 0.002/0.001, range = 0.1, rate2 = 1e-06
#> TH01: 10 alleles, mut model (M/F) = step-ext, rate = 0.002/0.001, range = 0.1, rate2 = 1e-06
#> D21S11: 26 alleles, mut model (M/F) = step-ext, rate = 0.002/0.001, range = 0.1, rate2 = 1e-06
#> D18S51: 23 alleles, mut model (M/F) = step-ext, rate = 0.002/0.001, range = 0.1, rate2 = 1e-06
#> PENTA_E: 21 alleles, mut model (M/F) = step-ext, rate = 0.002/0.001, range = 0.1, rate2 = 1e-06
#> D5S818: 9 alleles, mut model (M/F) = step-ext, rate = 0.002/0.001, range = 0.1, rate2 = 1e-06
#> D13S317: 9 alleles, mut model (M/F) = step-ext, rate = 0.002/0.001, range = 0.1, rate2 = 1e-06
#> D7S820: 19 alleles, mut model (M/F) = step-ext, rate = 0.002/0.001, range = 0.1, rate2 = 1e-06
#> D16S539: 9 alleles, mut model (M/F) = step-ext, rate = 0.002/0.001, range = 0.1, rate2 = 1e-06
#> CSF1PO: 11 alleles, mut model (M/F) = step-ext, rate = 0.002/0.001, range = 0.1, rate2 = 1e-06
#>
#> Converting to `ped` format
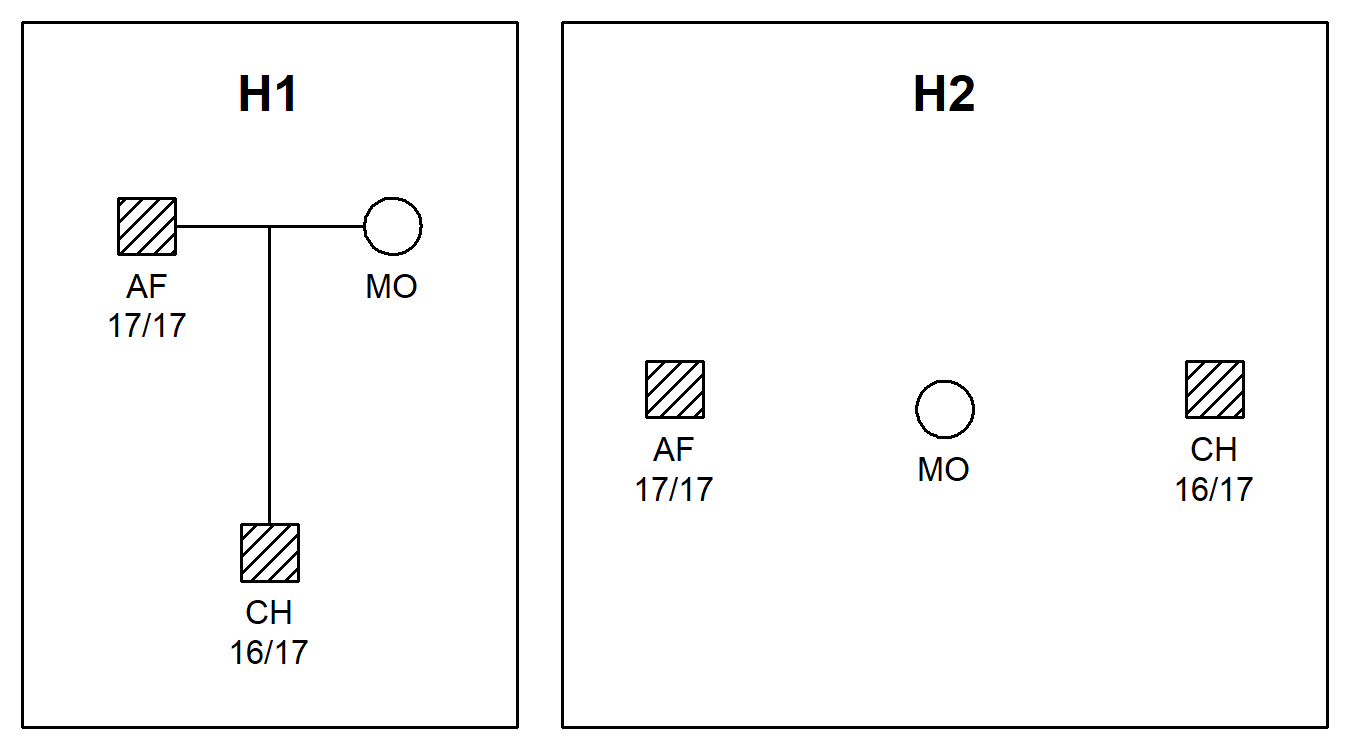
Here are the pedigrees, including genotypes for the first marker:
plotPedList(peds, hatched = typedMembers, marker = 1)
Further analysis of the data may be carried out with the forrel package. For instance, the following command computes the likelihood ratio comparing the two hypotheses:
forrel::kinshipLR(peds)
#> H1:H2 H2:H2
#> 760.3445 1.0000