Plant Photobiology Related Functions and Data.
photobiologyPlants 
Package ‘photobiologyPlants’ provides pre-defined functions for quantifying visible (‘VIS’), near infra-red (‘NIR’) and ultraviolet (‘UV’) radiation in relation to their effects on plants together with action spectra for photosynthesis. Absorbance spectra for the plant photoreceptors in families ‘phytochromes’, ‘cryptochromes’, ‘zeitlupe proteins’, ‘phototropins’ and ‘UVR8s’ are included together with absorbance spectra for plants’ “mass pigments”, including chlorophylls, carotenoids and some other metabolites such as flavonoids.
The package also includes data sets on the optical properties of plant organs, including whole-leaf reflectance, absorptance, transmittance and fluorescence spectra.
All data are derived from the scientific literature. Please, see the help pages for the different data sets for details about the primary sources of the data.
The package includes functions for the calculation of reference evapotranspiration using different approaches, for unit conversions for water in the atmosphere and for the computation of the energy balance of vegetation. These functions were included in package ‘photobiology’ (\< 0.12.0) and have been migrated to ‘photobiologyPlants’ (\>= 0.6.0).
The data in this package are stored in objects of classes defined in package ‘photobiology’ which are mostly backwards compatible with data frames but include metadata as attributes.
This package is part of a suite of R packages for photobiological calculations described at the r4photobiology web site.
Examples
library(photobiologyPlants)
eval_plots <- requireNamespace("ggspectra", quietly = TRUE)
if (eval_plots) library(ggspectra)
Spectral data are stored in R objects of classes defined in package ‘photobiology’.
class(McCree_photosynthesis.mspct$oats)
#> [1] "response_spct" "generic_spct" "tbl_df" "tbl"
#> [5] "data.frame"
Objects contain metadata that can be queried. The comment attribute commonly used in R.
comment(McCree_photosynthesis.mspct$oats)
#> [1] "One of the 'classical' action spectra of photosynthesis from K. J. McCree (1972): Avena sativa L. var. Coronado leaf sections."
And also other attributes defined in package ‘photobiology’.
what_measured(McCree_photosynthesis.mspct$oats)
#> [1] "Action spectrum of net CO2 uptake in Avena sativa L. var. Coronado (McCree 1972)."
how_measured(McCree_photosynthesis.mspct$oats)
#> [1] "Net CO2 uptake measured on detached leaf sections after about 2 to 10 minutes equilibration time at each wavelength"
is_normalised(McCree_photosynthesis.mspct$oats)
#> [1] TRUE
Functions defined in package ‘ggspectra’ make plotting easy. For example, to plot the action spectrum of photosynthesis in Oats we can use.
autoplot(McCree_photosynthesis.mspct$oats, unit.out = "photon")
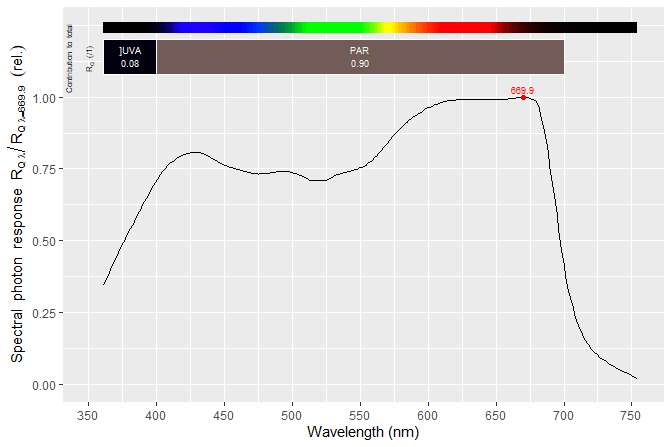
We can calculate the R:FR photon ratio of a light-source or iradiance spectrum, in this case the solar spectrum at ground level measured on at a specific location and time, included in package ‘photobiology’.
R_FR(sun.spct)
#> R:FR[q:q]
#> 1.266704
#> attr(,"radiation.unit")
#> [1] "q:q ratio"
We can also estimate the photo-equilibrium of phytochrome exposed in vitro to the same spectrum.
Pfr_Ptot(sun.spct)
#> [1] 0.68341
Installation
Installation of the most recent released version from CRAN (source and binaries available):
install.packages("photobiologyLamps")
Installation of the current unstable version from R-Universe CRAN-like repository (source and binaries available):
install.packages('photobiologySun',
repos = c('https://aphalo.r-universe.dev',
'https://cloud.r-project.org'))
Installation of the current unstable version from GitHub (only source available):
# install.packages("remotes")
remotes::install_github("aphalo/photobiologylamps")
Documentation
HTML documentation is available at (https://docs.r4photobiology.info/photobiologyPlants/), including the User Guide.
News on updates to the different packages of the ‘r4photobiology’ suite are regularly posted at (https://www.r4photobiology.info/).
Two articles introduce the basic ideas behind the design of the suite and describe its use: Aphalo P. J. (2015) (https://doi.org/10.19232/uv4pb.2015.1.14) and Aphalo P. J. (2016) (https://doi.org/10.19232/uv4pb.2016.1.15).
A book is under preparation, and the draft is currently available at (https://leanpub.com/r4photobiology/).
A handbook written before the suite was developed contains useful information on the quantification and manipulation of ultraviolet and visible radiation: Aphalo, P. J., Albert, A., Björn, L. O., McLeod, A. R., Robson, T. M., & Rosenqvist, E. (Eds.) (2012) Beyond the Visible: A handbook of best practice in plant UV photobiology (1st ed., p. xxx + 174). Helsinki: University of Helsinki, Department of Biosciences, Division of Plant Biology. ISBN 978-952-10-8363-1 (PDF), 978-952-10-8362-4 (paperback). PDF file available from (https://hdl.handle.net/10138/37558).
Contributing
Pull requests, bug reports, and feature requests are welcome at (https://github.com/aphalo/photobiologyPlants).
Citation
If you use this package to produce scientific or commercial publications, please cite according to:
citation("photobiologyPlants")
#> To cite package ‘photobiologyPlants’ in publications use:
#>
#> Aphalo, Pedro J. (2015) The r4photobiology suite. UV4Plants Bulletin,
#> 2015:1, 21-29. DOI:10.19232/uv4pb.2015.1.14
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Article{,
#> author = {Pedro J. Aphalo},
#> title = {The r4photobiology suite},
#> journal = {UV4Plants Bulletin},
#> volume = {2015},
#> number = {1},
#> pages = {21-29},
#> year = {2015},
#> doi = {10.19232/uv4pb.2015.1.14},
#> }
License
© 2015-2025 Pedro J. Aphalo ([email protected]). Released under the GPL, version 2 or greater. This software carries no warranty of any kind.

