Data for Sunlight Spectra.
photobiologySun 
Package ‘photobiologySun’ complements other packages in the R for photobiology suite. It contains spectral data for daylight, including sunlight and shade light. Package ‘photobiologySun’ also includes spectra measured or simulated for different times of the day, and reference spectra for solar radiation.
This package contains only data. Data are stored as collections of spectra of class source_mspct from package ‘photobiology’, which is the core of the R for photobiology suite. Spectra can be easily plotted with functions and methods from package ‘ggspectra’. The spectra can be used seamlessly with functions from package ‘photobioloy’. However, class source_mspct is derived from list and class source_spct is derived from data.frame making the data also usable as is with base R functions.
Examples
library(ggspectra)
library(photobiologySun)
How many spectra are contained in the collection of spectra gap.mspct? In this case the spectra are from a fast time series of measurements.
length(gap.mspct)
#> [1] 72
What are their names?
names(gap.mspct)
#> [1] "spct.01" "spct.02" "spct.03" "spct.04" "spct.05" "spct.06" "spct.07"
#> [8] "spct.08" "spct.09" "spct.10" "spct.11" "spct.12" "spct.13" "spct.14"
#> [15] "spct.15" "spct.16" "spct.17" "spct.18" "spct.19" "spct.20" "spct.21"
#> [22] "spct.22" "spct.23" "spct.24" "spct.25" "spct.26" "spct.27" "spct.28"
#> [29] "spct.29" "spct.30" "spct.31" "spct.32" "spct.33" "spct.34" "spct.35"
#> [36] "spct.36" "spct.37" "spct.38" "spct.39" "spct.40" "spct.41" "spct.42"
#> [43] "spct.43" "spct.44" "spct.45" "spct.46" "spct.47" "spct.48" "spct.49"
#> [50] "spct.50" "spct.51" "spct.52" "spct.53" "spct.54" "spct.55" "spct.56"
#> [57] "spct.57" "spct.58" "spct.59" "spct.60" "spct.61" "spct.62" "spct.63"
#> [64] "spct.64" "spct.65" "spct.66" "spct.67" "spct.68" "spct.69" "spct.70"
#> [71] "spct.71" "spct.72"
How many spectra are contained in the collection of spectra sun_reference.mspct?
length(sun_reference.mspct)
#> [1] 5
What are their names? (The three “AM0” spectra are for the extraterrestrial solar spectrum, AM0 = zero atmosphera. ASTM G173 corresponds to AM1.5 and frequently used in engineering.)
names(sun_reference.mspct)
#> [1] "ASTM.E490.AM0" "ASTM.G173.direct" "ASTM.G173.global" "Gueymard.AM0"
#> [5] "WMO.Wehrli.AM0"
Summary calculations can be easily done with methods from package ‘photobiology’. Here we calculate photon irradiance with function q_irrad() for another collection of spectra. As the spectra are normalised we pass allow.scaled = TRUE,
q_irrad(sun_hourly_august.spct, scale.factor = 1e6) # Q_Total: umol m-2 s-1
#> # A tibble: 31 × 3
#> Q_Total spct.idx when.measured
#> <dbl> <chr> <dttm>
#> 1 98.9 spct.01 2014-08-21 03:30:00
#> 2 169. spct.02 2014-08-21 04:30:00
#> 3 564. spct.03 2014-08-21 05:30:00
#> 4 780. spct.04 2014-08-21 06:30:00
#> 5 534. spct.05 2014-08-21 07:30:00
#> 6 1255. spct.06 2014-08-21 08:30:00
#> 7 1889. spct.07 2014-08-21 09:30:00
#> 8 1914. spct.08 2014-08-21 10:30:00
#> 9 1482. spct.09 2014-08-21 11:30:00
#> 10 1264. spct.10 2014-08-21 12:30:00
#> # ℹ 21 more rows
The autoplot() methods from package ‘ggspectra’ can be used for plotting one or more spectra at a time.
autoplot(gap.mspct[1:10], annotations = c("-", "peaks")) +
theme_bw()
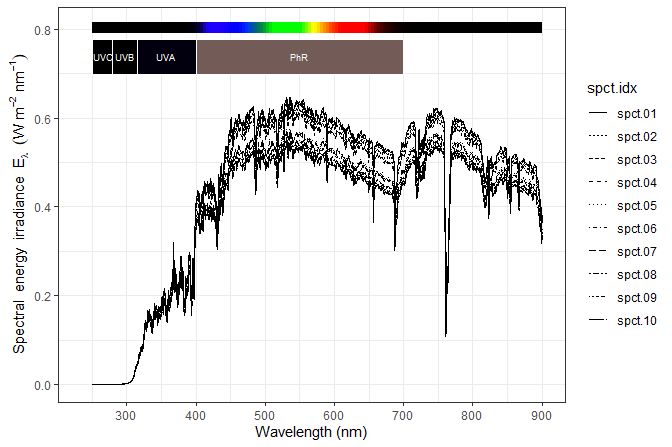
The classes of the objects used to store the spectral data are derived from "data.frame" making direct use of the data with functions and methods from base R and various packages easy.
Installation
Installation of the most recent stable version from CRAN (source and binaries available):
install.packages("photobiologySun")
Installation of the current unstable version from R-Universe CRAN-like repository (source and binaries available):
install.packages('photobiologySun',
repos = c('https://aphalo.r-universe.dev',
'https://cloud.r-project.org'))
Installation of the current unstable version from GitHub (only source available):
# install.packages("devtools")
devtools::install_github("aphalo/photobiologySun")
Documentation
HTML documentation is available at (https://docs.r4photobiology.info/photobiologySun/), including a User Guide.
News on updates to the different packages of the ‘r4photobiology’ suite are regularly posted at (https://www.r4photobiology.info/).
Two articles introduce the basic ideas behind the design of the suite and its use: Aphalo P. J. (2015) (https://doi.org/10.19232/uv4pb.2015.1.14) and Aphalo P. J. (2016) (https://doi.org/10.19232/uv4pb.2016.1.15).
A book is under preparation, and the draft is currently available at (https://leanpub.com/r4photobiology/).
A handbook written before the suite was developed contains useful information on the quantification and manipulation of ultraviolet and visible radiation: Aphalo, P. J., Albert, A., Björn, L. O., McLeod, A. R., Robson, T. M., & Rosenqvist, E. (Eds.) (2012) Beyond the Visible: A handbook of best practice in plant UV photobiology (1st ed., p. xxx + 174). Helsinki: University of Helsinki, Department of Biosciences, Division of Plant Biology. ISBN 978-952-10-8363-1 (PDF), 978-952-10-8362-4 (paperback). PDF file available from (https://hdl.handle.net/10138/37558).
Contributing
Pull requests, bug reports, and feature requests are welcome at (https://github.com/aphalo/photobiologySun).
Citation
If you use this package to produce scientific or commercial publications, please cite according to:
citation("photobiologySun")
#> To cite package ‘photobiologySun’ in publications use:
#>
#> Aphalo, Pedro J. (2015) The r4photobiology suite. UV4Plants Bulletin,
#> 2015:1, 21-29. DOI:10.19232/uv4pb.2015.1.14
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Article{,
#> author = {Pedro J. Aphalo},
#> title = {The r4photobiology suite},
#> journal = {UV4Plants Bulletin},
#> volume = {2015},
#> number = {1},
#> pages = {21-29},
#> year = {2015},
#> doi = {10.19232/uv4pb.2015.1.14},
#> }
License
© 2012-2024 Pedro J. Aphalo ([email protected]). Released under the GPL, version 2 or greater. This software carries no warranty of any kind.

