Prediction Intervals for Random Forests.
piRF - Prediction Intervals for Random Forests
Chancellor Johnstone and Haozhe Zhang
Introduction
The goal of piRF is to implement multiple state-of-the art random forest prediction interval methodologies in one complete package. Currently, the methods implemented can only be utilized within isolated packages, or the authors have not made a package publicly available. The package itself utilizes the functionality provided by the ranger package.
Installation
You can install the released version of piRF from CRAN with:
install.packages("piRF")
And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("chancejohnstone/piRF")
Example
This is a basic example which utilizes the airfoil dataset included with piRF. The dataset comes from UCI Archive. The NASA data set comprises different size NACA 0012 airfoils at various wind tunnel speeds and angles of attack.
The follwoing functions are not exported by piRF but are used for this example.
library(piRF)
## basic example code
data(airfoil)
head(airfoil)
#> freq AoA length velocity disp pressure
#> 1 800 0 0.3048 71.3 0.00266337 126.201
#> 2 1000 0 0.3048 71.3 0.00266337 125.201
#> 3 1250 0 0.3048 71.3 0.00266337 125.951
#> 4 1600 0 0.3048 71.3 0.00266337 127.591
#> 5 2000 0 0.3048 71.3 0.00266337 127.461
#> 6 2500 0 0.3048 71.3 0.00266337 125.571
#functions to get average length and average coverage of output
getPILength <- function(x){
#average PI length across each set of predictions
l <- x[,2] - x[,1]
avg_l <- mean(l)
return(avg_l)
}
getCoverage <- function(x, response){
#output coverage for test data
coverage <- sum((response >= x[,1]) * (response <= x[,2]))/length(response)
return(coverage)
}
Prediction intervals are generated for each of the methods implemented using train and test datasets constructed from the airfoil data.
method_vec <- c("quantile", "Zhang", "Tung", "Romano", "Roy", "HDI", "Ghosal")
#generate train and test data
set.seed(2020)
ratio <- .975
nrow <- nrow(airfoil)
n <- floor(nrow*ratio)
samp <- sample(1:nrow, size = n)
train <- airfoil[samp,]
test <- airfoil[-samp,]
#generate prediction intervals
res <- rfint(pressure ~ . , train_data = train, test_data = test,
method = method_vec,
concise= FALSE,
num_threads = 2)
In this example, the num_threads option identifies the use of two cores for parallel processing. The default is to use all available cores. The concise option allows for the output of predictions for the test observations.
Below are the coverage rates and average prediction interval lengths using the test dataset. Both are important characteristics of prediction intervals.
#empirical coverage, and average prediction interval length for each method
coverage <- sapply(res$int, FUN = getCoverage, response = test$pressure)
coverage
#> quantile Zhang Tung Romano Roy HDI Ghosal
#> 0.8947368 0.8947368 0.9210526 0.9210526 0.9473684 0.9473684 0.9736842
length <- sapply(res$int, FUN = getPILength)
length
#> quantile Zhang Tung Romano Roy HDI Ghosal
#> 10.233720 7.035656 7.810820 10.578972 11.789397 9.895526 9.097531
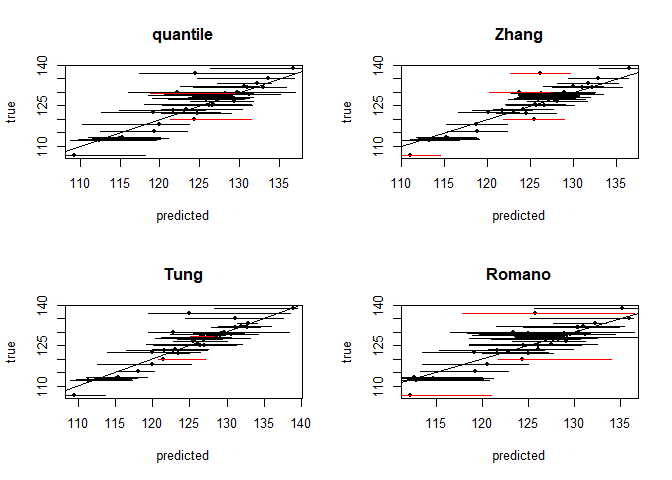
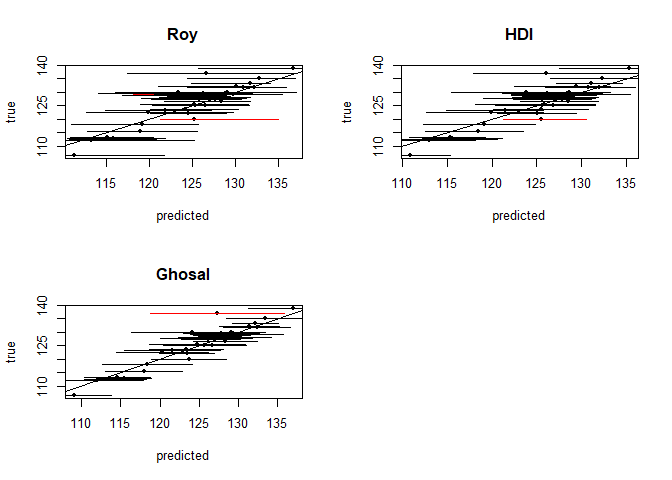
Below are plots of the resulting prediction intervals generated for each method.
#plotting intervals and predictions
par(mfrow = c(2,2))
for(i in 1:7){
col <- ((test$pressure >= res$int[[i]][,1]) *
(test$pressure <= res$int[[i]][,2])-1)*(-1)+1
plot(x = res$preds[[i]], y = test$pressure, pch = 20,
col = "black", ylab = "true", xlab = "predicted", main = method_vec[i])
abline(a = 0, b = 1)
segments(x0 = res$int[[i]][,1], x1 = res$int[[i]][,2],
y1 = test$pressure, y0 = test$pressure, lwd = 1, col = col)
}
If you find any issues with the package, or have suggestions for improvements, please let us know.
References
Breiman, Leo. 2001. “Random Forests.” Machine Learning 45 (1). Springer: 5–32. https://link.springer.com/article/10.1023/A:1010933404324.
Ghosal, Indrayudh, and Giles Hooker. 2018. “Boosting Random Forests to Reduce Bias; One-Step Boosted Forest and Its Variance Estimate.” ArXiv Preprint. https://arxiv.org/pdf/1803.08000.pdf.
Meinshausen, Nicolai. 2006. “Quantile Regression Forests.” Journal of Machine Learning Research 7 (Jun): 983–99. http://www.jmlr.org/papers/volume7/meinshausen06a/meinshausen06a.pdf.
Romano, Yaniv, Evan Patterson, and Emmanuel Candes. 2019. “Conformalized Quantile Regression.” ArXiv Preprint. https://arxiv.org/pdf/1905.03222v1.pdf.
Roy, Marie-Hélène, and Denis Larocque. 2019. “Prediction Intervals with Random Forests.” Statistical Methods in Medical Research. SAGE Publications Sage UK: London, England. https://doi.org/10.1177/0962280219829885.
Tung, Nguyen Thanh, Joshua Zhexue Huang, Thuy Thi Nguyen, and Imran Khan. 2014. “Bias-Corrected Quantile Regression Forests for High-Dimensional Data.” In 2014 International Conference on Machine Learning and Cybernetics, 1:1–6. IEEE. https://link.springer.com/article/10.1007/s10994-014-5452-1.
Zhang, Haozhe, Joshua Zimmerman, Dan Nettleton, and Daniel J. Nordman. 2019. “Random Forest Prediction Intervals.” The American Statistician. Taylor & Francis, 1–15. https://doi.org/10.1080/00031305.2019.1585288.
Zhu, Lin, Jiaxin Lu, and Yihong Chen. 2019. “HDI-Forest: Highest Density Interval Regression Forest.” ArXiv Preprint. https://arxiv.org/pdf/1905.10101.pdf.