Useful Functions for Visualization.
plutor
In ancient Roman mythology,
Plutowas the ruler of the underworld and presides over the afterlife.Plutowas frequently conflated withPlutus, the god of wealth, because mineral wealth was found underground.When plotting with R, you try once, twice, practice again and again, and finally you get a pretty figure you want.
It’s a
plot tour, a tour about repetition and reward.Hope
plutorhelps you on the tour!
installation
You can install the development version of plutor like so:
devtools::install_github("william-swl/plutor")
And load the package:
library(plutor)
It is recommended to perform initialization, which adjusts the default plotting parameters in an interactive environment (such as jupyter notebook) and sets the default theme to theme_pl().
pl_init()
plots
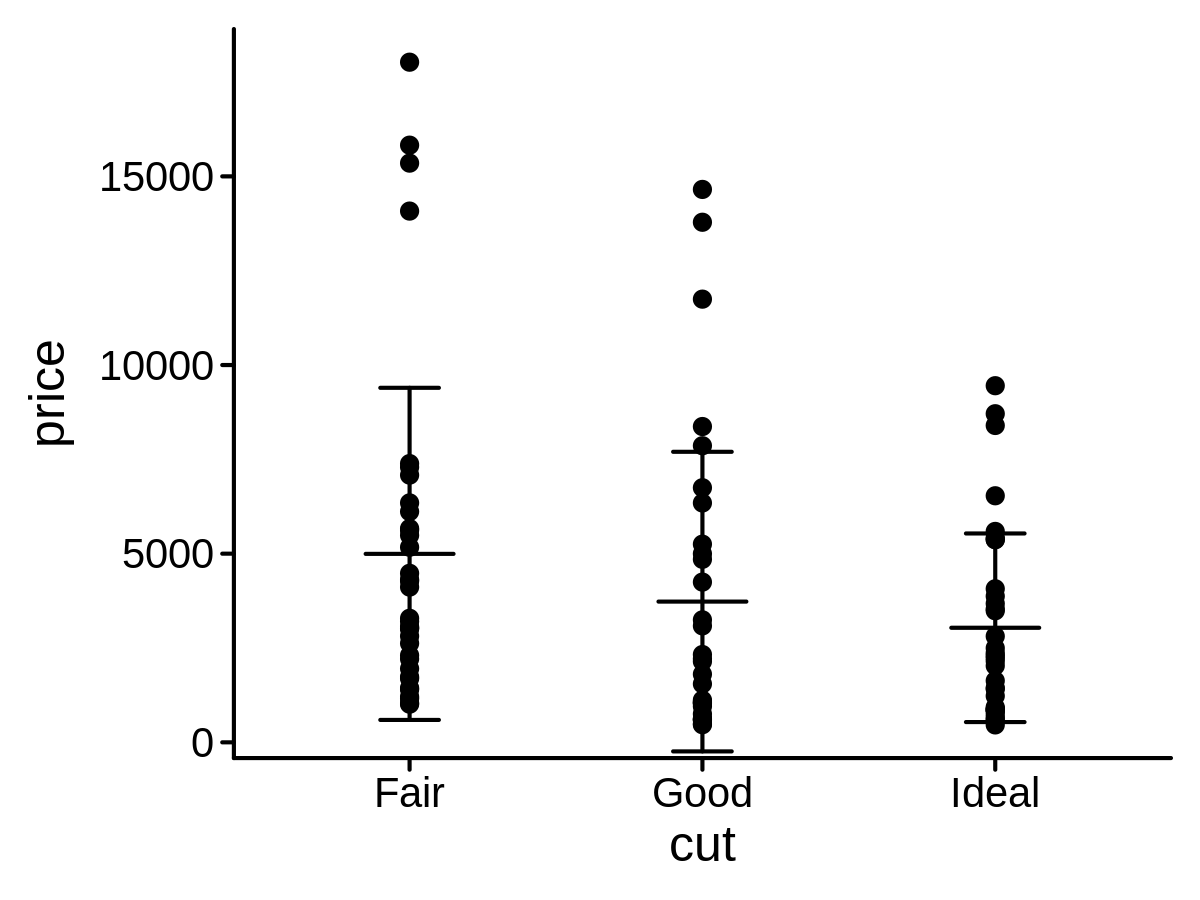
Description values plot:
The describe geom is used to create description values plot, including center symbol and error symbol.
The center symbol can be mean, median or other custom functions.
The error symbol can be sd, quantile or other custom functions.
mini_diamond %>% ggplot(aes(x = cut, y = price)) +
geom_point() +
geom_describe()
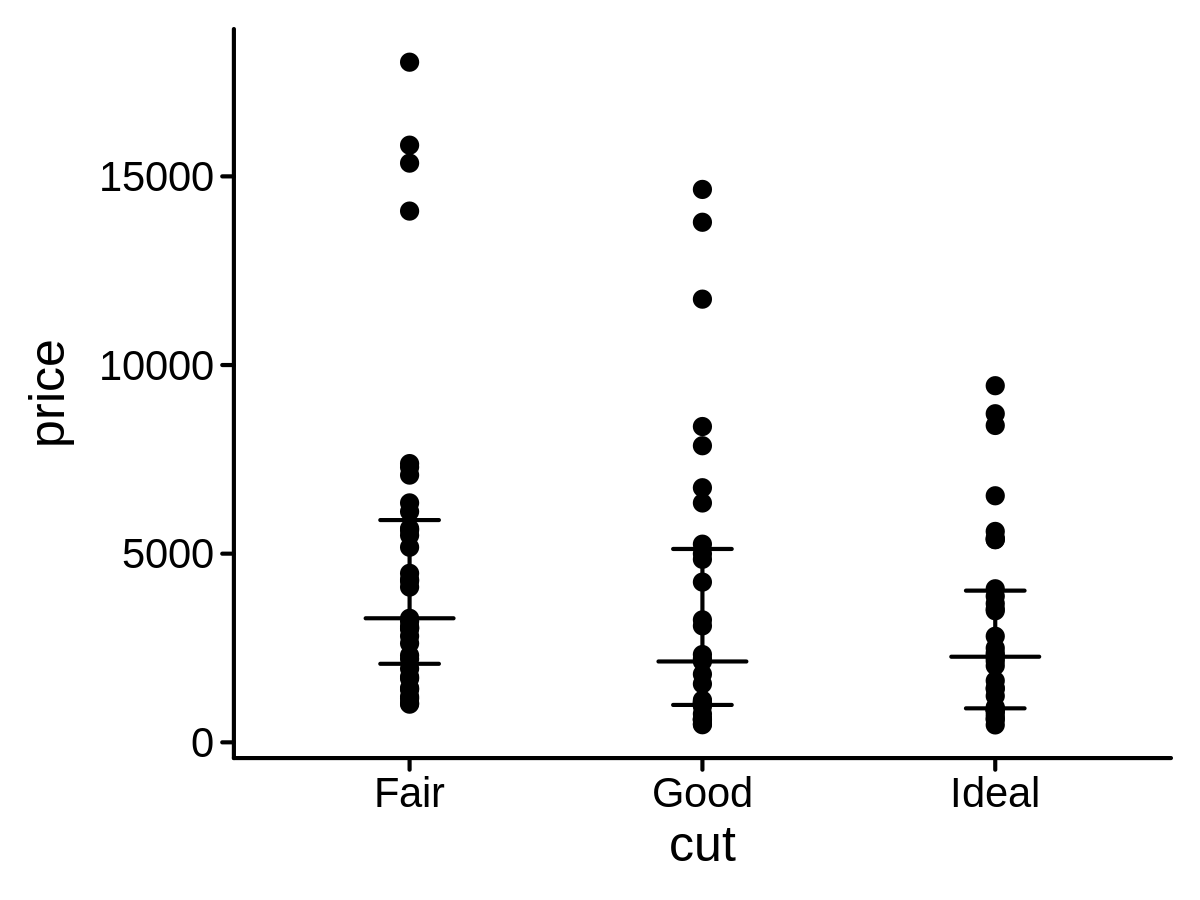
center_func <- median
low_func <- function(x, na.rm) {
quantile(x, 0.25, na.rm = na.rm)
}
high_func <- function(x, na.rm) {
quantile(x, 0.75, na.rm = na.rm)
}
mini_diamond %>% ggplot(aes(x = cut, y = price)) +
geom_point() +
geom_describe(center_func = center_func, low_func = low_func, high_func = high_func)
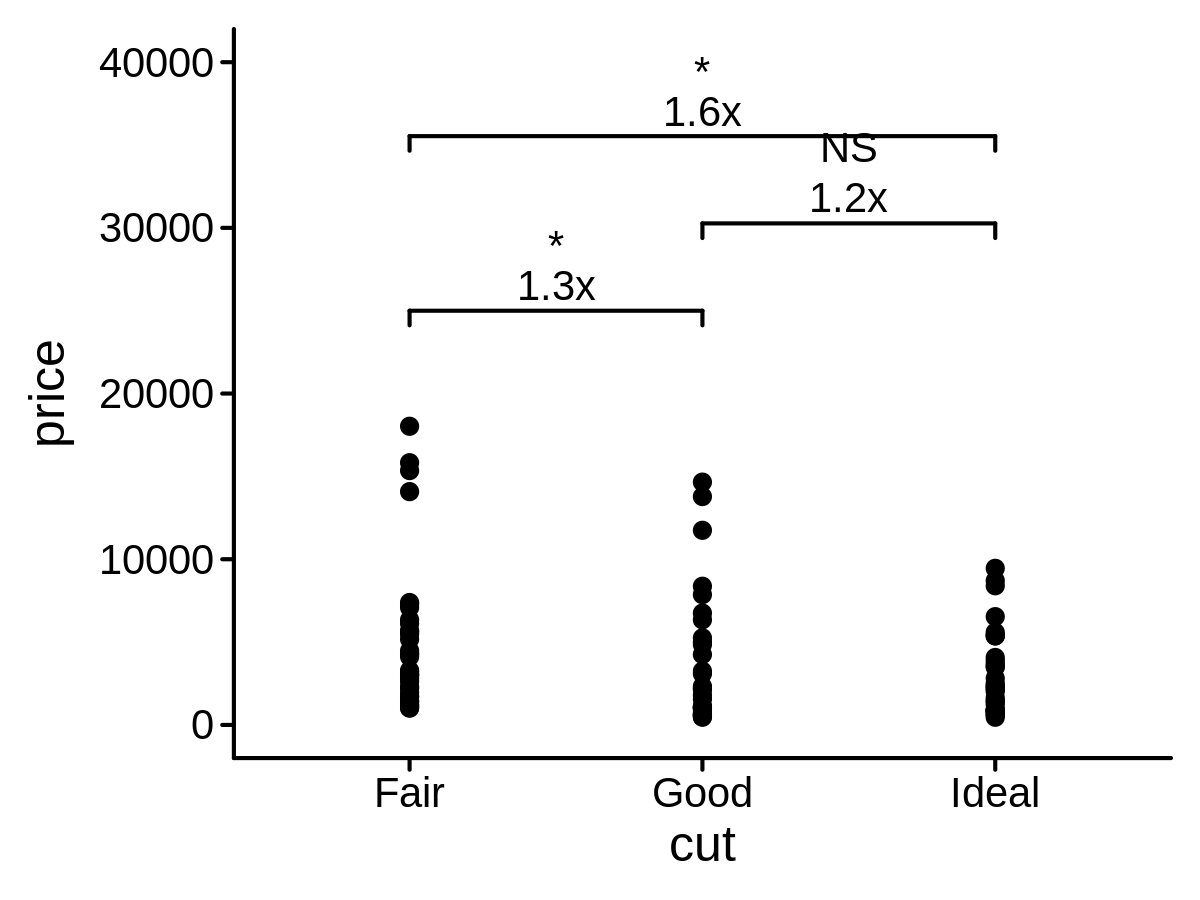
Add p value and fold change on a plot
p <- ggplot(data = mini_diamond, mapping = aes(x = cut, y = price)) +
geom_point() +
geom_compare(cp_label = c("psymbol", "right_deno_fc"), lab_pos = 25000, step_increase = 0.3) +
ylim(0, 40000)
p
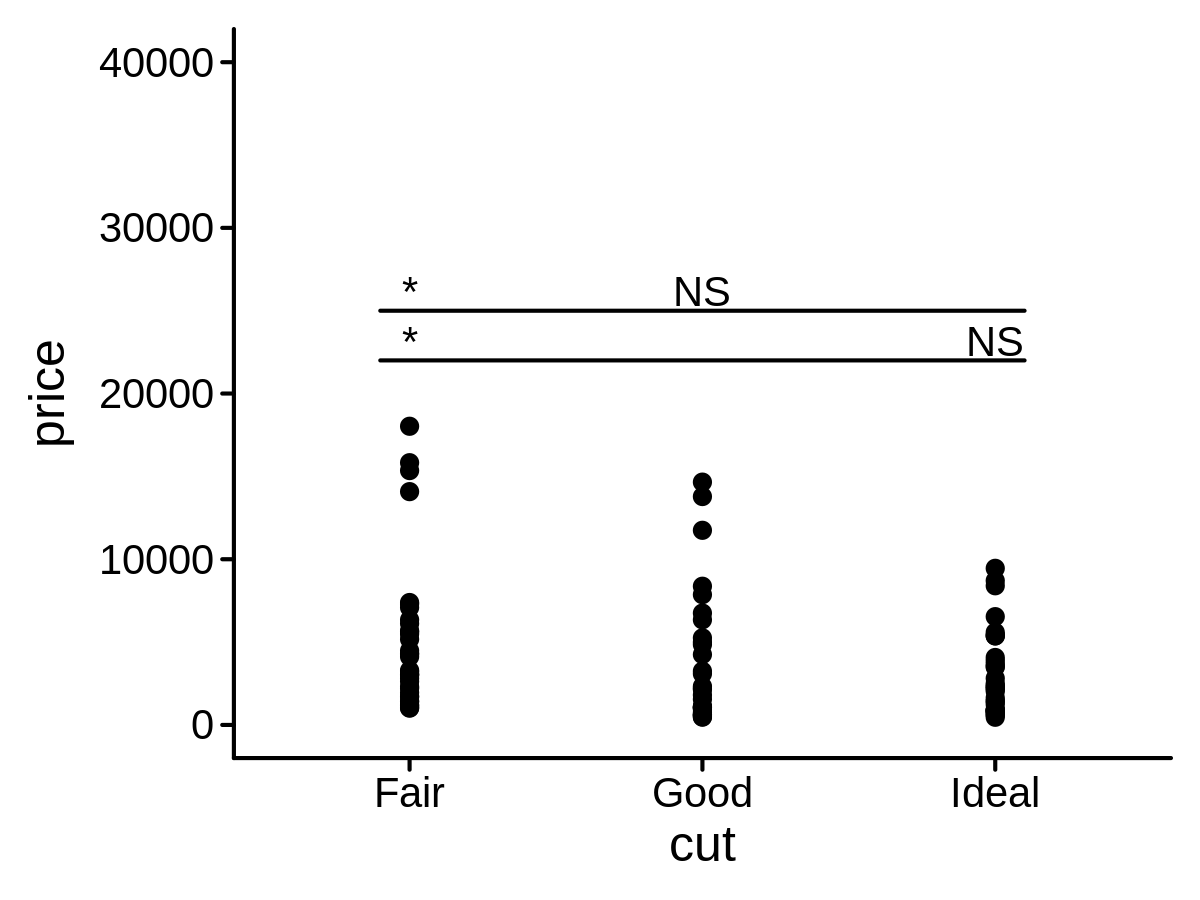
ggplot(data = mini_diamond, mapping = aes(x = cut, y = price)) +
geom_point() +
geom_compare(cp_ref = "Good", cp_inline = TRUE, lab_pos = 22000, brackets_widen = 0.1) +
geom_compare(cp_ref = "Ideal", cp_inline = TRUE, lab_pos = 25000, brackets_widen = 0.1) +
ylim(0, 40000)
extract the result of geom_compare from a ggplot object
head(extract_compare(p))
#> PANEL x xend n1 n2 p plim psymbol y1 y2 fc
#> 1 1 1 2 35 31 0.041 0.05 * 4995.057 3730.387 1.339018
#> 2 1 2 3 31 34 0.93 1.01 NS 3730.387 3036.588 1.228480
#> 3 1 1 3 35 34 0.018 0.05 * 4995.057 3036.588 1.644957
#> right_deno_fc left_deno_fc label cp_step y yend group
#> 1 1.3x 0.75x *\n1.3x 0 25000.0 25000.0 1
#> 2 1.2x 0.81x NS\n1.2x 1 30269.2 30269.2 1
#> 3 1.6x 0.61x *\n1.6x 2 35538.4 35538.4 1
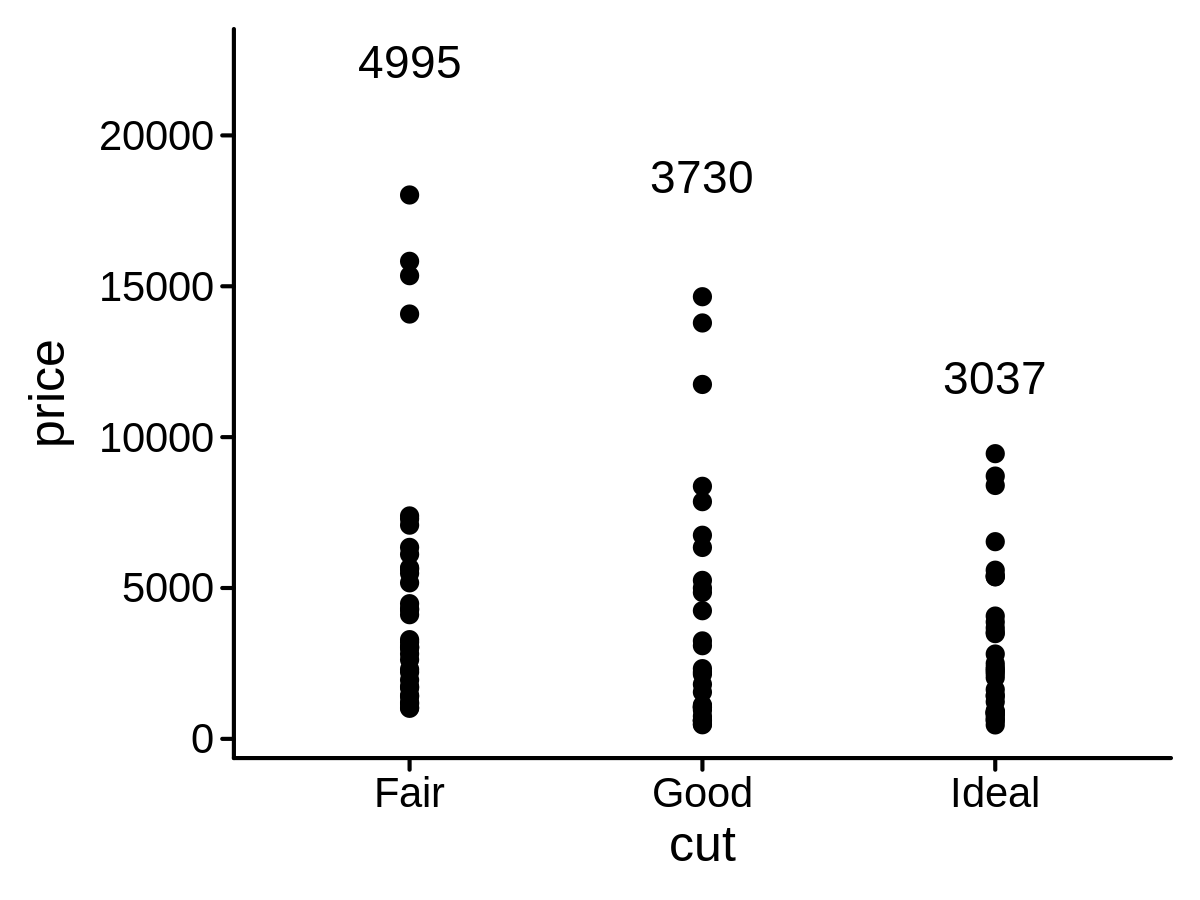
A new Stat class to add mean labels on a plot
mini_diamond %>% ggplot(aes(x = cut, y = price)) +
geom_point() +
geom_text(aes(label = price), stat = "meanPL")
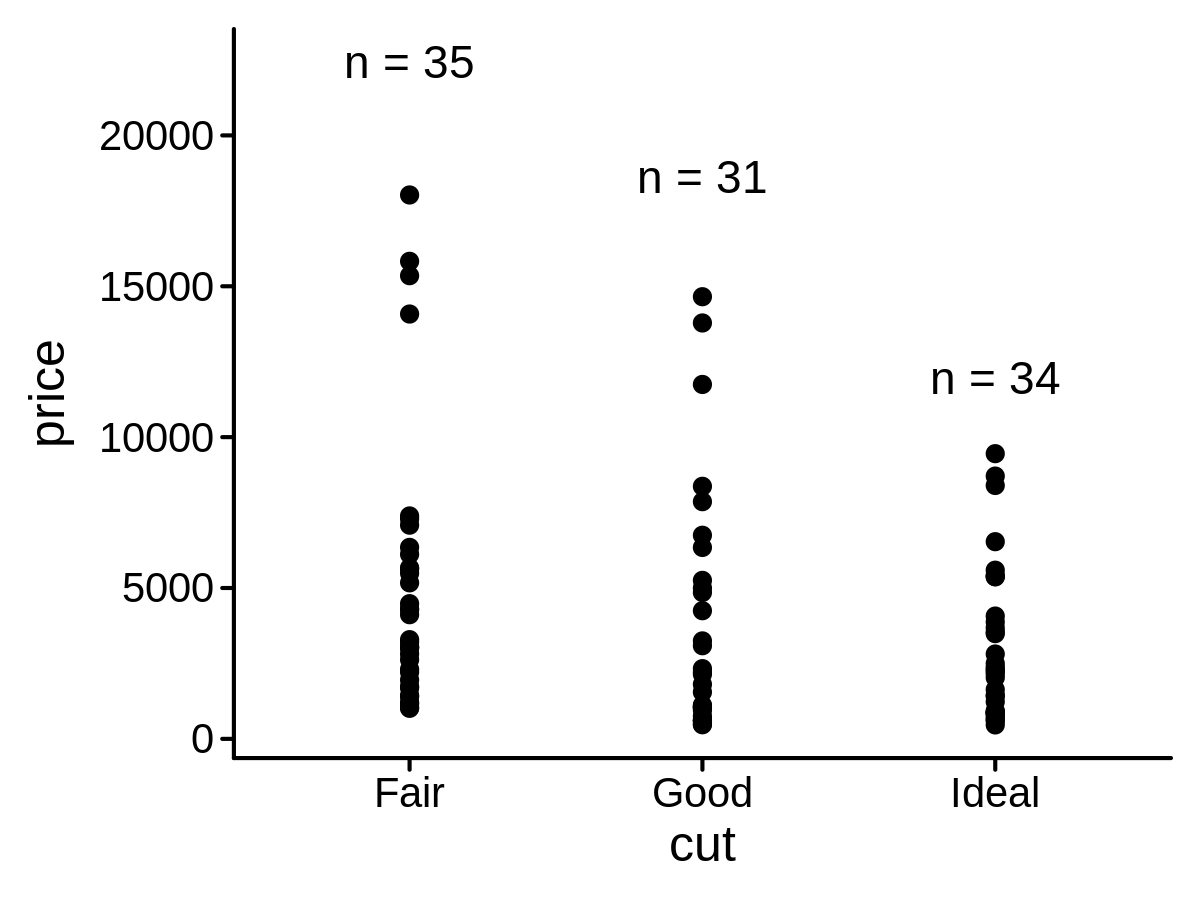
A new Stat class to add count labels on a plot
mini_diamond %>% ggplot(aes(x = cut, y = price)) +
geom_point() +
geom_text(aes(label = price), stat = "countPL")
A new Stat class to add custom function labels on a plot
lab_func <- function(x) {
str_glue("mean = {round(mean(x))}\nn = {length(x)}")
}
mini_diamond %>% ggplot(aes(y = cut, x = price)) +
geom_point() +
geom_text(aes(label = price),
stat = "funcPL",
lab_func = lab_func, lab_pos = 25000
) +
xlim(0, 30000)
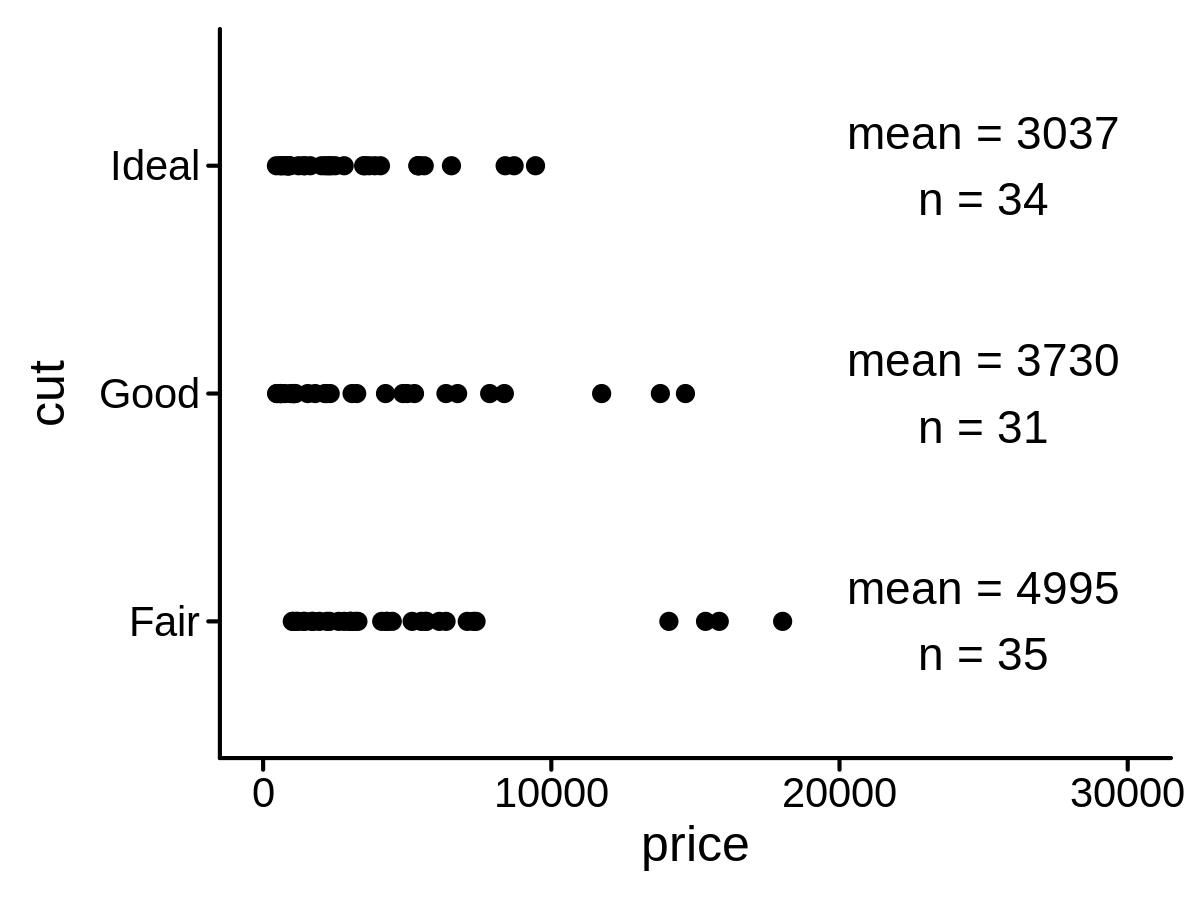
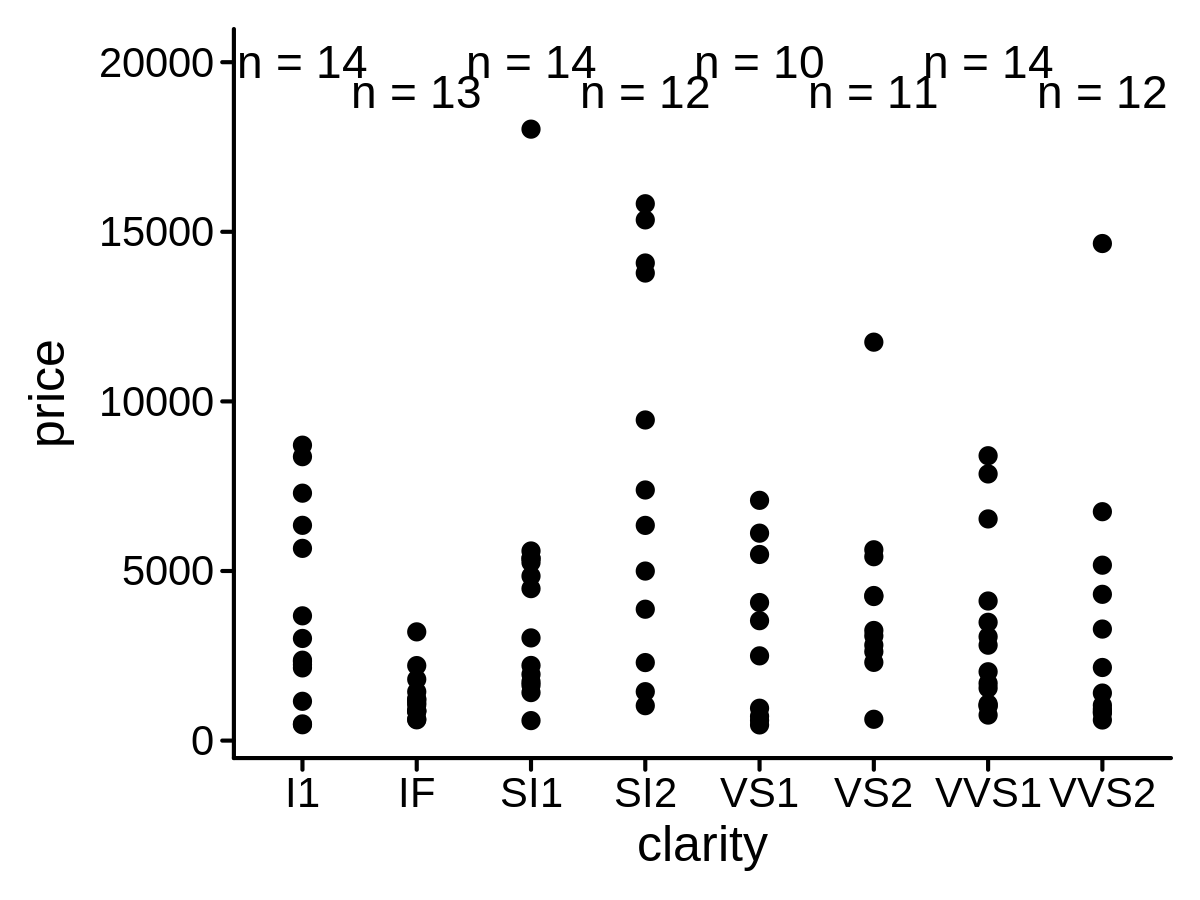
A new Position function to create float x/y position
mini_diamond %>% ggplot(aes(x = clarity, y = price)) +
geom_point() +
geom_text(aes(label = price),
stat = "countPL",
lab_pos = 20000, position = position_floatyPL()
)
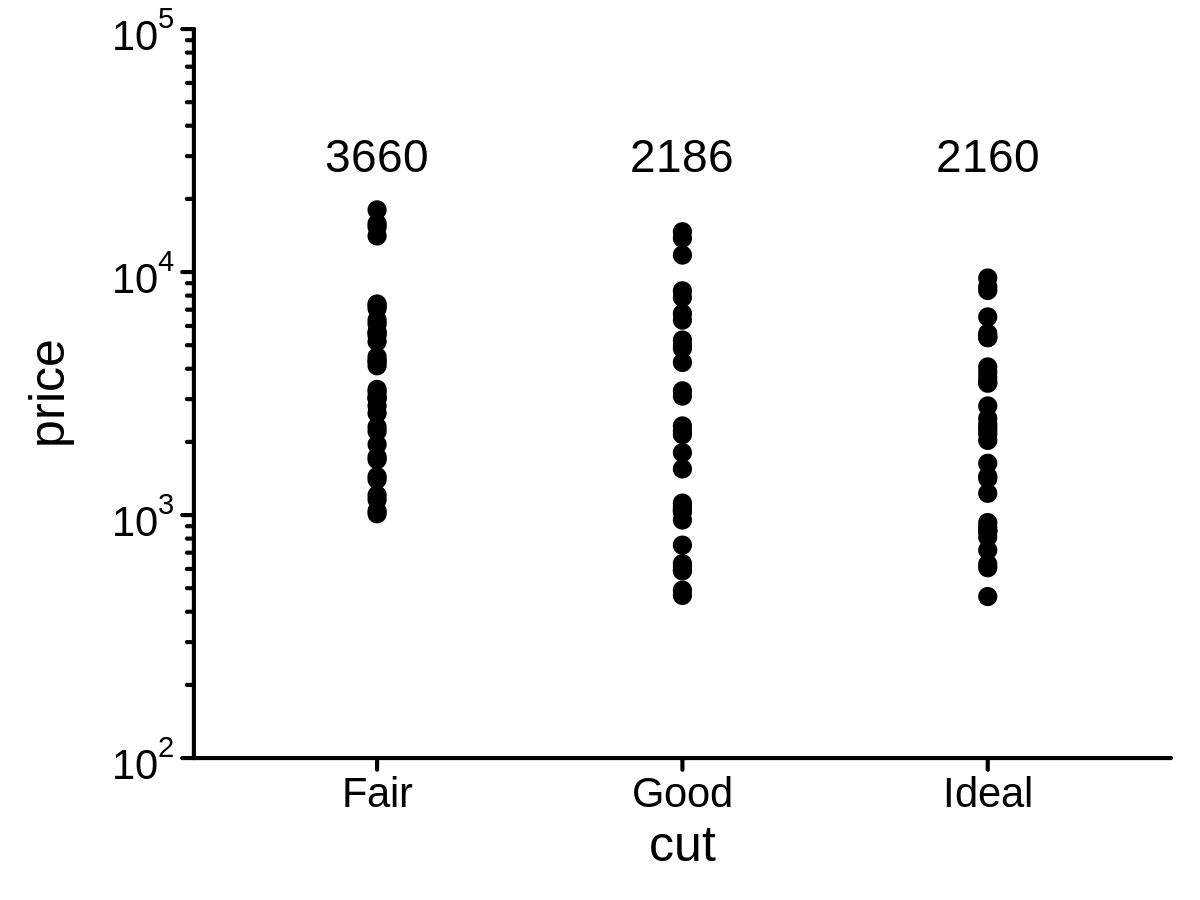
A variant of scale_y_log10() to show axis minor breaks and better axis labels
mini_diamond %>% ggplot(aes(x = cut, y = price)) +
geom_point() +
geom_text(stat = "meanPL", lab_pos = 30000) +
scale_y_log10_pl(show_minor_breaks = TRUE, limits = c(100, 100000))
A variant of scale_y_continuous() to show axis minor breaks
mini_diamond %>% ggplot(aes(x = cut, y = price)) +
geom_point() +
geom_text(stat = "meanPL", lab_pos = 25000) +
scale_y_continuous_pl(limits = c(0, 40000), minor_break_step = 2500)
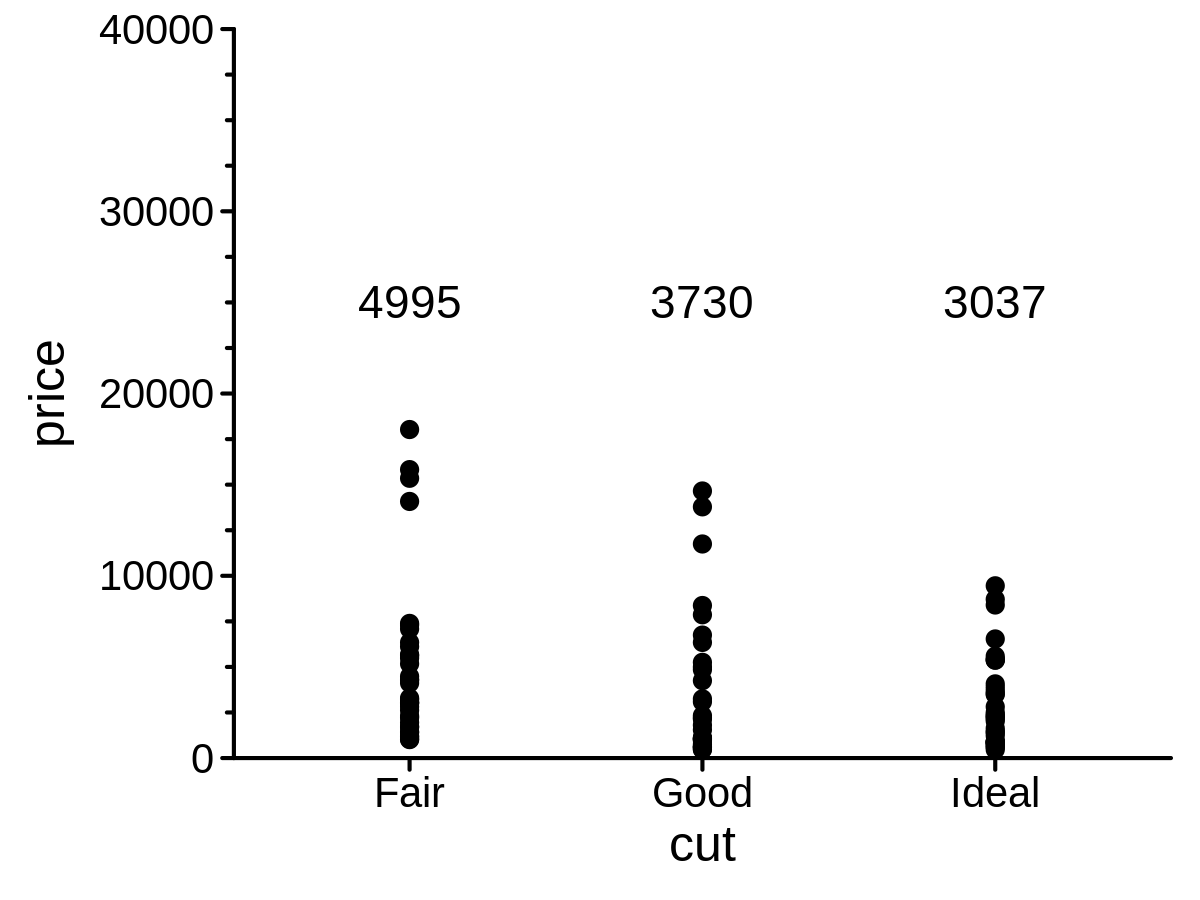
color
- generate gradient colors
gradient_colors(c("blue", "red"), 10)
#> [1] "#0000FF" "#1C00E2" "#3800C6" "#5500AA" "#71008D" "#8D0071" "#AA0055"
#> [8] "#C60038" "#E2001C" "#FF0000"
- show colors
plot_colors(gradient_colors(c("blue", "red"), 10))

- select colors from
RColorBrewerpackage presets
brewer_colors("Blues", 5) %>% plot_colors()

- select colors from
ggscipackage presets
sci_colors("npg", 5) %>% plot_colors()

- assign colors by a column in a tibble, for the convenience to use
scale_color_identity()
assign_colors(mini_diamond, cut, colors = sci_colors("nejm", 8))
#> # A tibble: 100 × 8
#> id carat cut clarity price x y assigned_colors
#> <chr> <dbl> <chr> <chr> <int> <dbl> <dbl> <chr>
#> 1 id-1 1.02 Fair SI1 3027 6.25 6.18 #BC3C29FF
#> 2 id-2 1.51 Good VS2 11746 7.27 7.18 #0072B5FF
#> 3 id-3 0.52 Ideal VVS1 2029 5.15 5.18 #E18727FF
#> 4 id-4 1.54 Ideal SI2 9452 7.43 7.45 #E18727FF
#> 5 id-5 0.72 Ideal VS1 2498 5.73 5.77 #E18727FF
#> 6 id-6 2.02 Fair SI2 14080 8.33 8.37 #BC3C29FF
#> 7 id-7 0.27 Good VVS1 752 4.1 4.07 #0072B5FF
#> 8 id-8 0.51 Good SI2 1029 5.05 5.08 #0072B5FF
#> 9 id-9 1.01 Ideal SI1 5590 6.43 6.4 #E18727FF
#> 10 id-10 0.7 Fair VVS1 1691 5.56 5.41 #BC3C29FF
#> # … with 90 more rows
- colors of nucleotides and amino acids
# bioletter_colors
theme
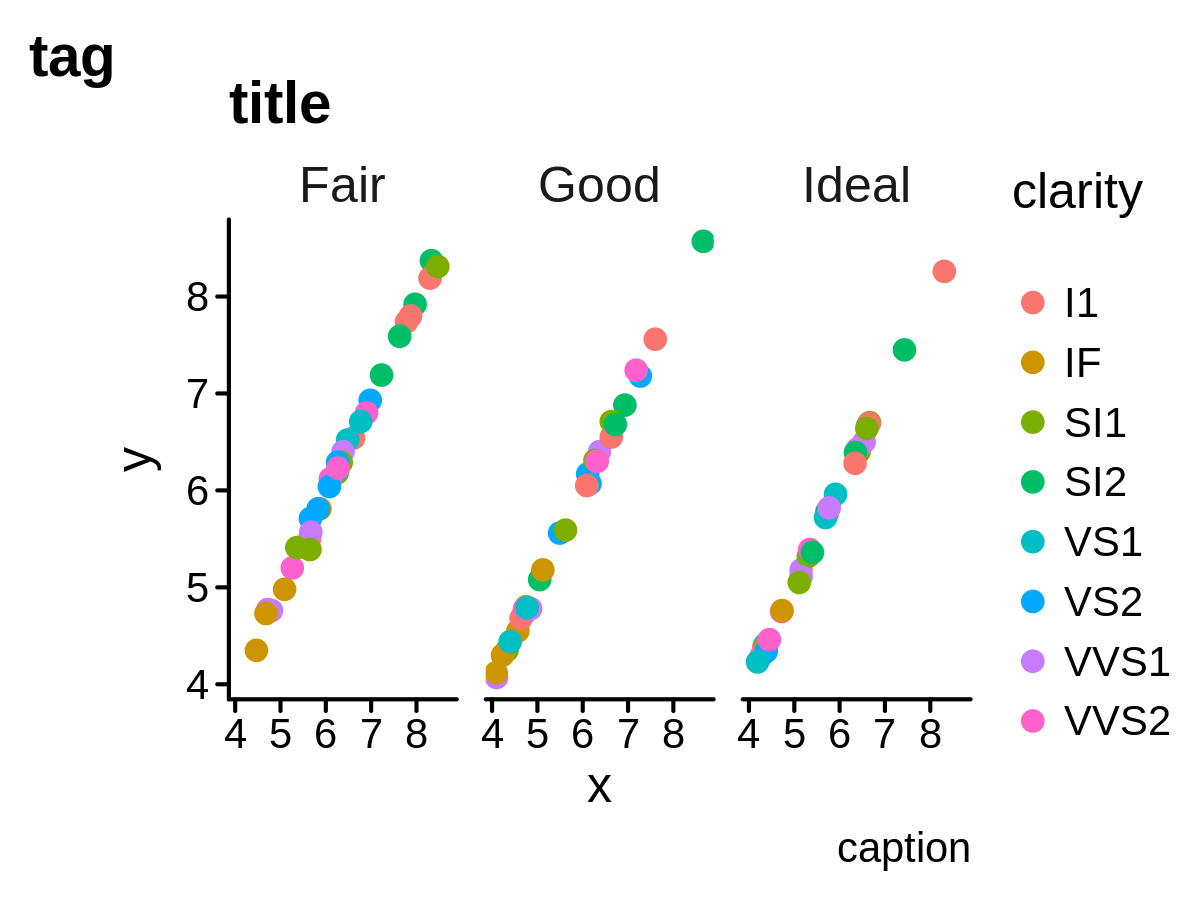
- a custom flexible theme
ggplot(mini_diamond, aes(x = x, y = y, color = clarity)) +
geom_point(size = 2) +
facet_grid(. ~ cut) +
labs(title = "title", tag = "tag", caption = "caption") +
theme_pl()
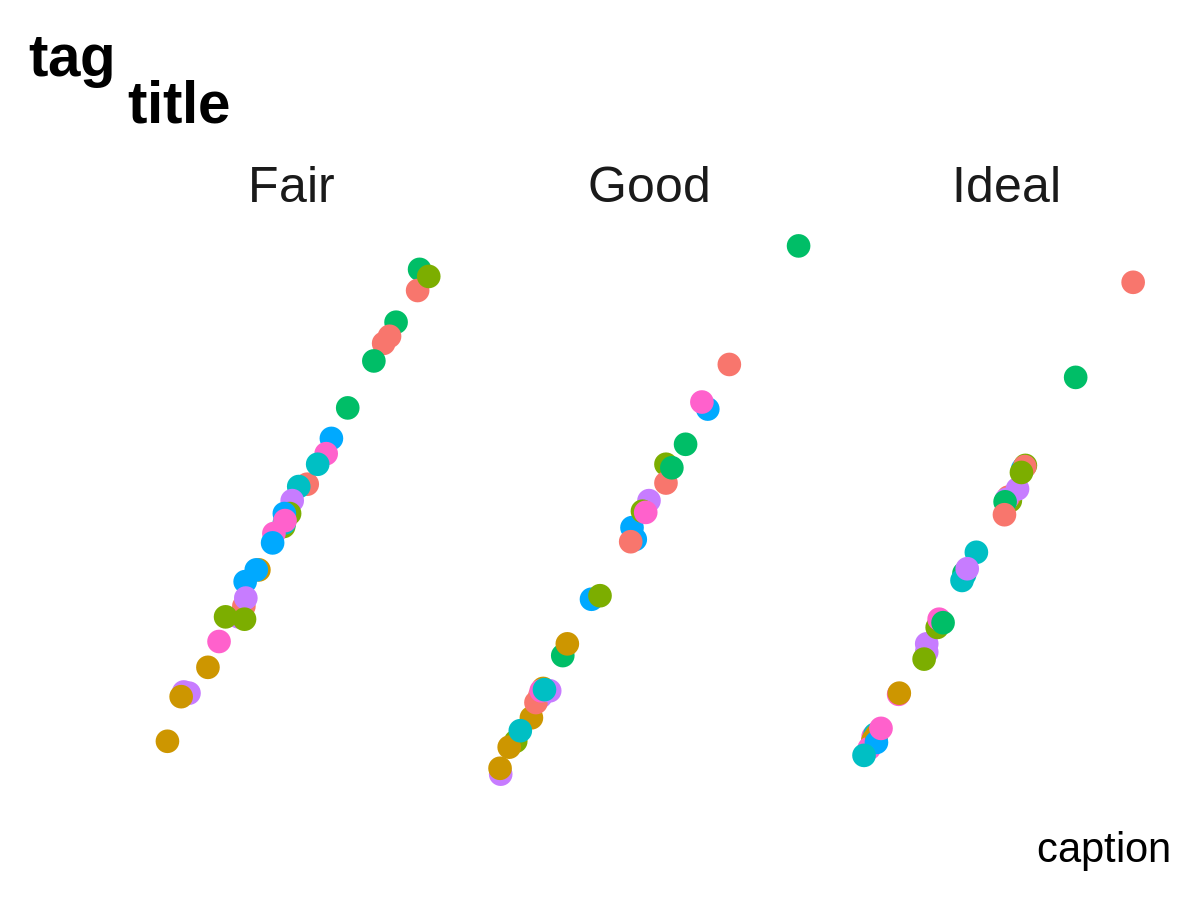
- a blank theme
ggplot(mini_diamond, aes(x = x, y = y, color = clarity)) +
geom_point(size = 2) +
facet_grid(. ~ cut) +
labs(title = "title", tag = "tag", caption = "caption") +
theme_pl0()
- a fixed mapping from size in
geom_xxxto unitptunder 300 dpi
# for text and points
# geom_point(..., size = ppt(5))
# geom_text(..., size = tpt(5))
# for lines
# geom_line(..., linewidth = lpt(1))
- set repr size and resolution
pl_size(w = 4, h = 3, res = 300)
- units transformation
# inches <-> centimeters
inch2cm(1)
#> [1] 2.54
#> attr(,"unit")
#> [1] 1
in2cm(1)
#> [1] 2.54
#> attr(,"unit")
#> [1] 1
cm2inch(1)
#> [1] 0.3937008
#> attr(,"unit")
#> [1] 2
cm2in(1)
#> [1] 0.3937008
#> attr(,"unit")
#> [1] 2
# inches <-> millimeters
inch2mm(1)
#> [1] 25.4
#> attr(,"unit")
#> [1] 7
in2mm(1)
#> [1] 25.4
#> attr(,"unit")
#> [1] 7
mm2inch(1)
#> [1] 0.03937008
#> attr(,"unit")
#> [1] 2
mm2in(1)
#> [1] 0.03937008
#> attr(,"unit")
#> [1] 2
# points <-> centimeters
pt2cm(1)
#> [1] 0.03514598
#> attr(,"unit")
#> [1] 1
cm2pt(1)
#> [1] 28.45276
#> attr(,"unit")
#> [1] 8
# points <-> millimeters
pt2mm(1)
#> [1] 0.3514598
#> attr(,"unit")
#> [1] 7
mm2pt(1)
#> [1] 2.845276
#> attr(,"unit")
#> [1] 8
IO
- save a plot
# pl_save(p, 'plot.pdf', width=14, height=10)
- save a plot into an blank A4 canvas, or a custom canvas
# pl_save(p, 'plot.pdf', width=14, height=10, canvas='A4', units='cm')
# pl_save(p, 'plot.pdf', width=14, height=10, canvas=c(20, 25), units='cm')
