Description
Graphical Check for Proportional Odds Assumption.
Description
Implements the method described at the UCLA Statistical Consulting site <https://stats.idre.ucla.edu/r/dae/ordinal-logistic-regression/> for checking if the proportional odds assumption holds for a cumulative logit model.
README.md
pomcheckr
The goal of pomcheckr is to implement the method described by UCLA Statistical Consulting for checking if the proportional odds assumption holds for a cumulative logit model.
Installation
You can install the released version of pomcheckr from CRAN with:
install.packages("pomcheckr")
You can install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("melissa-wong/pomcheckr")
Examples
Default Syntax
The following example uses the default syntax:
library(pomcheckr)
plot(pomcheck("Species", c("Sepal.Length", "Sepal.Width"),
iris))
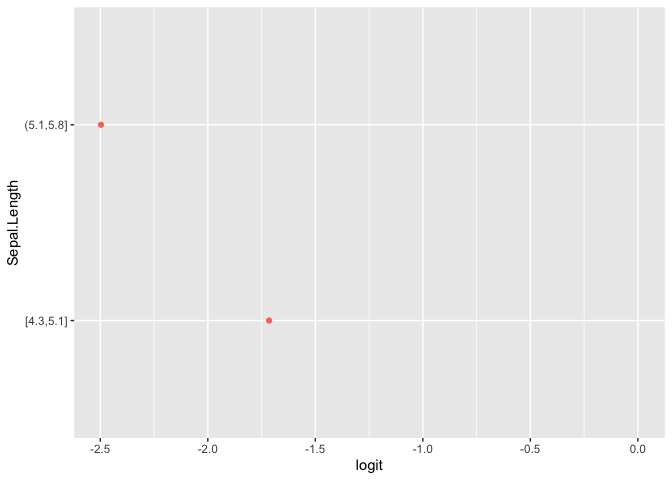
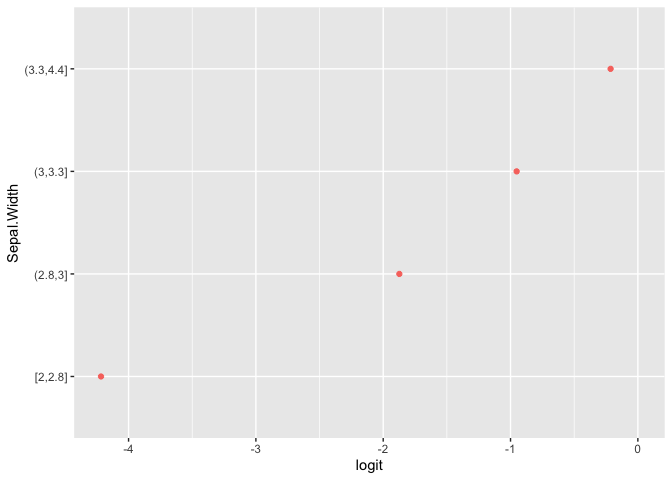
Formula Syntax
This is the equivalent example using formula syntax:
plot(pomcheck(Species ~ Sepal.Length + Sepal.Width, iris))
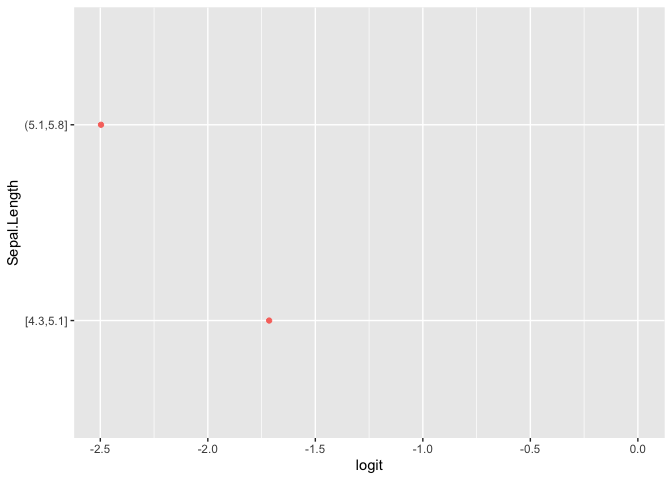
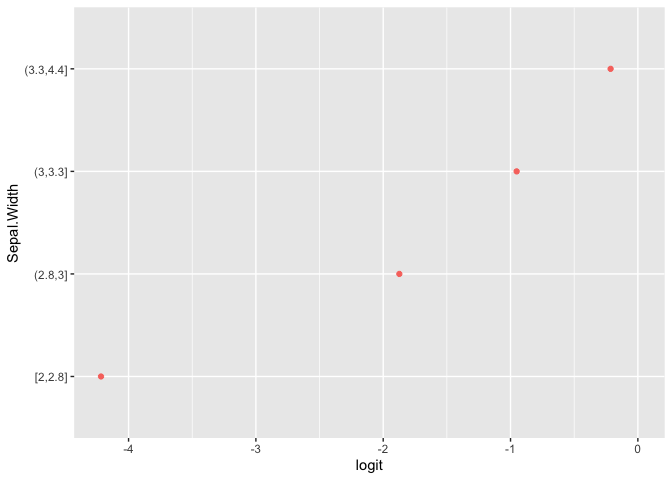
Examine Data
This example shows how to access and view the pomcheck object underlying the plots:
p <- pomcheck(Species ~ Sepal.Length + Sepal.Width, iris)
summary(p)
#> Length Class Mode
#> [1,] 4 grouped_df list
#> [2,] 4 grouped_df list
print(p)
#> [[1]]
#> # A tibble: 4 x 4
#> # Groups: Sepal.Length [4]
#> Sepal.Length `Species_>=1` `Species_>=2` `Species_>=3`
#> <fct> <dbl> <dbl> <dbl>
#> 1 [4.3,5.1] Inf -1.97 -3.69
#> 2 (5.1,5.8] Inf 0.580 -1.92
#> 3 (5.8,6.4] NA Inf 0.0572
#> 4 (6.4,7.9] NA Inf 1.06
#>
#> [[2]]
#> # A tibble: 4 x 4
#> # Groups: Sepal.Width [4]
#> Sepal.Width `Species_>=1` `Species_>=2` `Species_>=3`
#> <fct> <dbl> <dbl> <dbl>
#> 1 [2,2.8] Inf 3.83 -0.388
#> 2 (2.8,3] Inf 1.42 -0.452
#> 3 (3,3.3] Inf 0.547 -0.405
#> 4 (3.3,4.4] Inf -1.64 -1.86
#>
#> attr(,"class")
#> [1] "pomcheck" "list"