Description
Creates a Prisma Diagram using 'DiagrammeR'.
Description
Creates 'PRISMA' <http://prisma-statement.org/> diagram from a minimal dataset of included and excluded studies and allows for more custom diagrams. 'PRISMA' diagrams are used to track the identification, screening, eligibility, and inclusion of studies in a systematic review.
README.md
prismadiagramR
The goal of prismadiagramR is to create a custom prismadiagram in R.
Installation
You can install the released version of prismadiagramR from CRAN with:
install.packages("prismadiagramR")
And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("ltrainstg/prismadiagramR")
Example
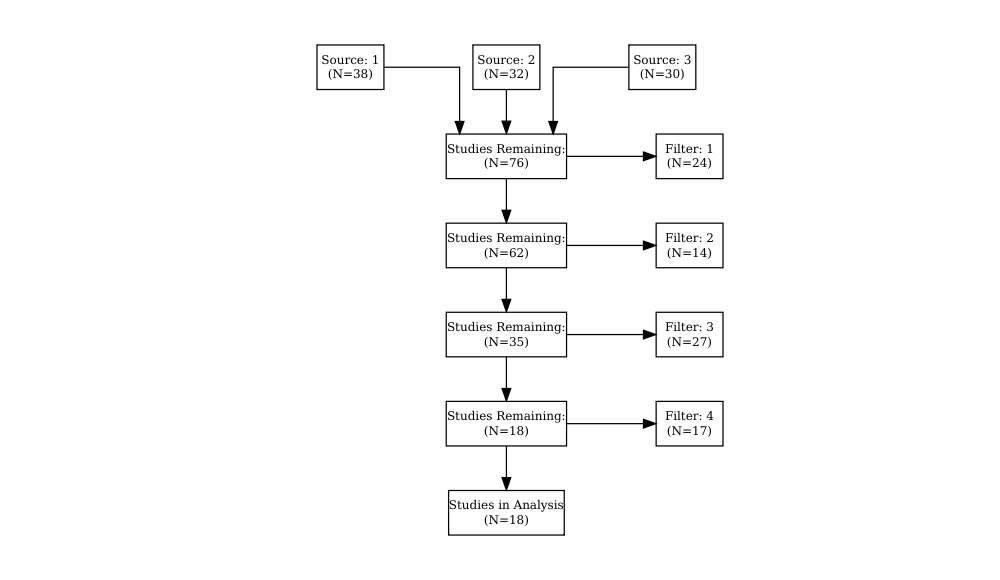
This example shows how to create a simple automated PRISMA from a publication tracker.
set.seed(25)
N <- 100
studyStatus <- data.frame(Pub.ID = seq(1:N),
Source = sample(1:3, N, replace = TRUE),
Filter = sample(1:5, N, replace = TRUE))
studyStatus$Filter[studyStatus$Filter==5] <- NA
getPrisma(studyStatus) %>% DiagrammeR::grViz(.)
#> Warning in getPrisma(studyStatus): prismaFormat is null so attempting to
#> make automatic one from studyStatus
#> Warning in getFormatNode(prismaFormat): fontSize param not passed in
#> prismaFormat
Other PRISMA resources
A few other R packages exist that also make PRISMA diagram that might be better for your needs.
- prismaStatement This also uses DiagrammeR, but the template is fixed.
- metagear This does not use DiagrammeR and is highly customizable, but is buried with many other functions and a little hard to get working.
Package Development Resources
This package was developed following these guides:
- John Muschelli’s r package development 1 HERE
- https://sahirbhatnagar.com/rpkg/#tests.
