Creates and Plots P-Value Functions, S-Value Functions, Confidence Distributions and Confidence De….
pvaluefunctions
P-value functions 
Accompanying paper
We published an accompanying paper to illustrate the use of p-value functions:
Infanger D, Schmidt-Trucksäss A. (2019): P value functions: An underused method to present research results and to promote quantitative reasoning. Statistics in Medicine.38: 4189-4197. doi: 10.1002/sim.8293.
Recreation of the figures in the paper
The code and instructions to reproduce all graphics in our paper can be found in the following GitHub repository: https://github.com/DInfanger/pvalue_functions
Overview
This is the repository for the R-package pvaluefunctions. The package contains R functions to create graphics of p-value functions, confidence distributions, confidence densities, or the Surprisal value (S-value) (Greenland 2019).
Installation
You can install the package directly from CRAN by typing install.packages("pvaluefunctions"). After installation, load it in R using library(pvaluefunctions).
Dependencies
The function depends on the following R packages, which need to be installed beforehand:
Use the command install.packages(c("ggplot2", "scales", "zipfR", "pracma", "gsl")) in R to install those packages.
Examples
For more examples and code, see the vignette.
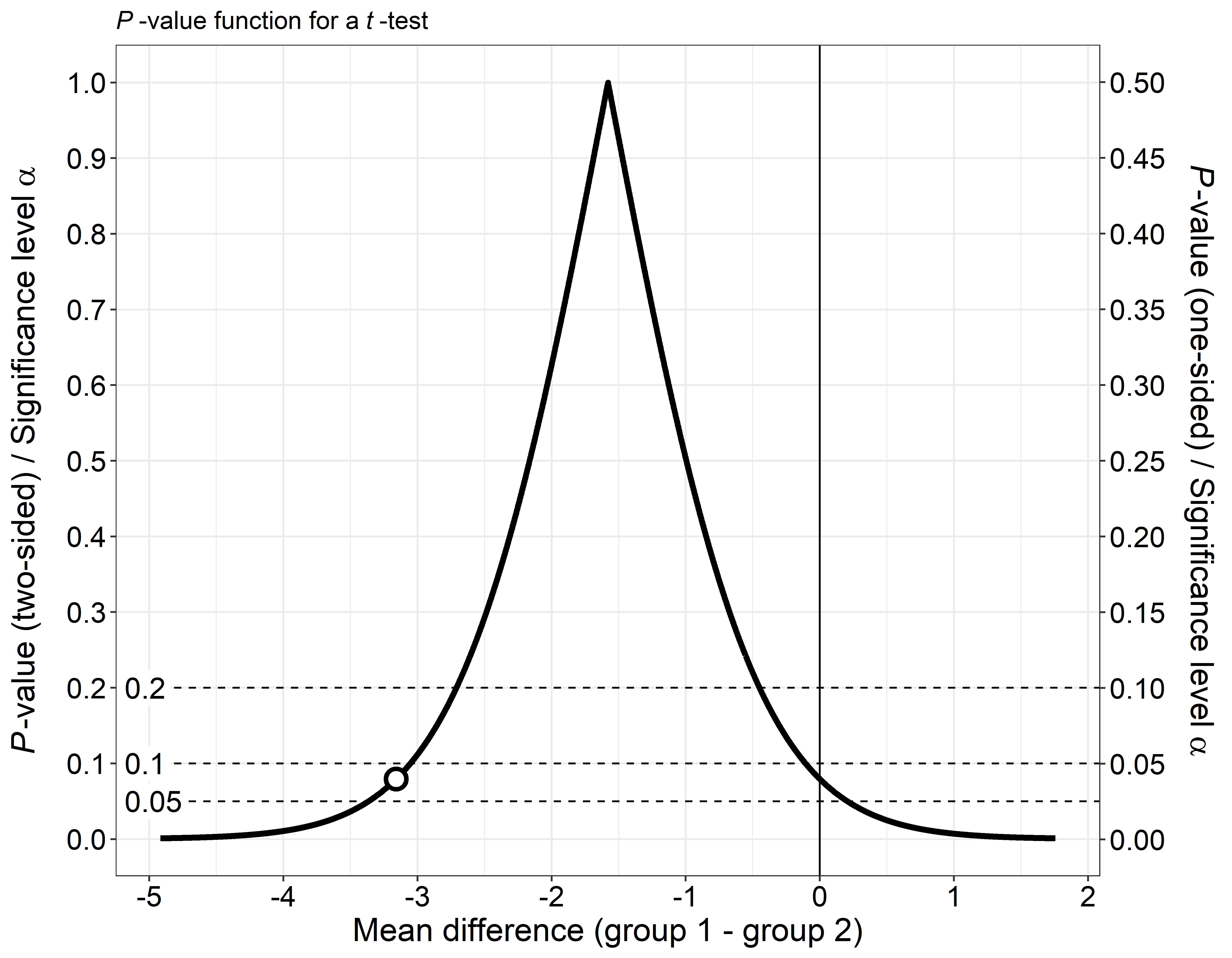
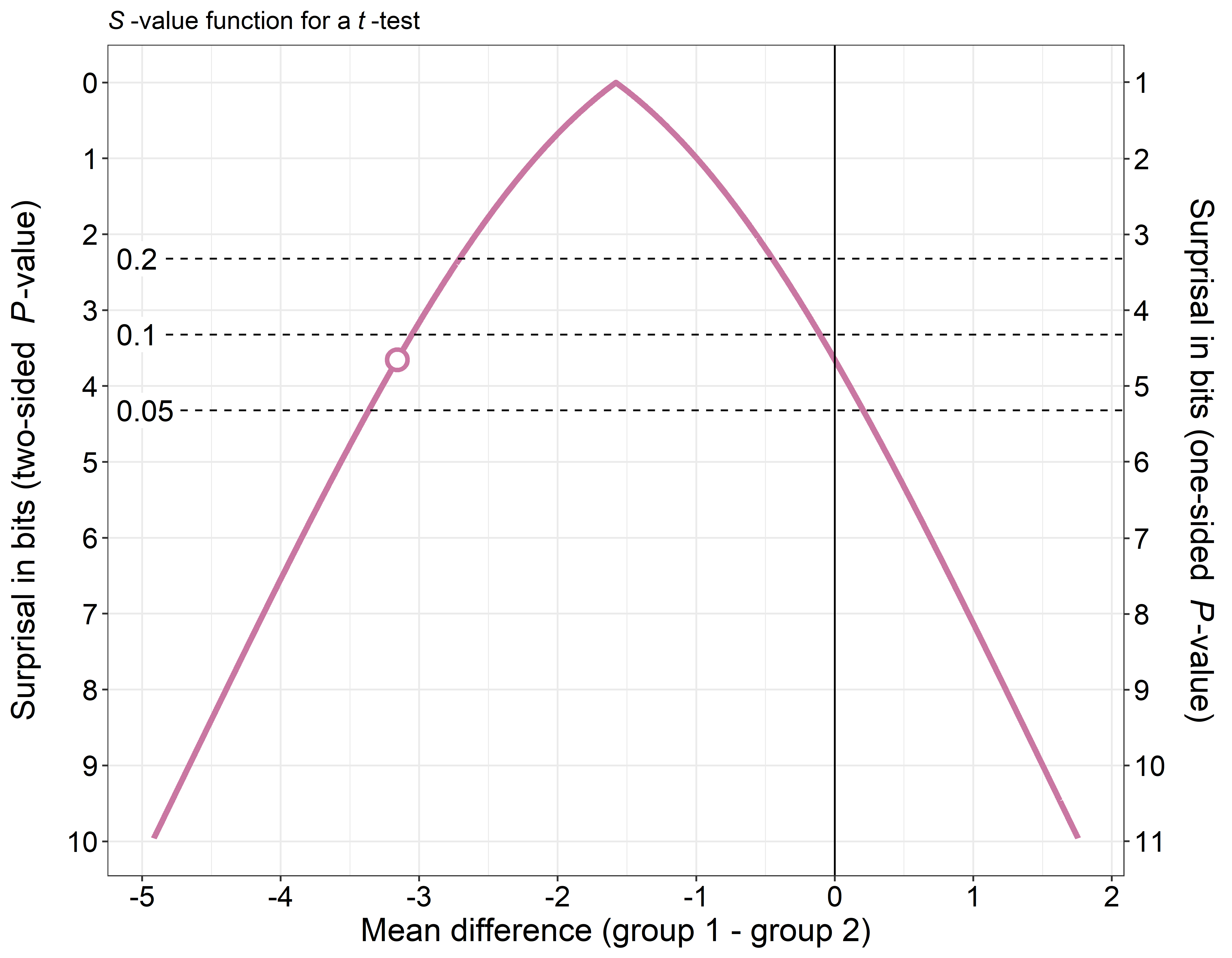
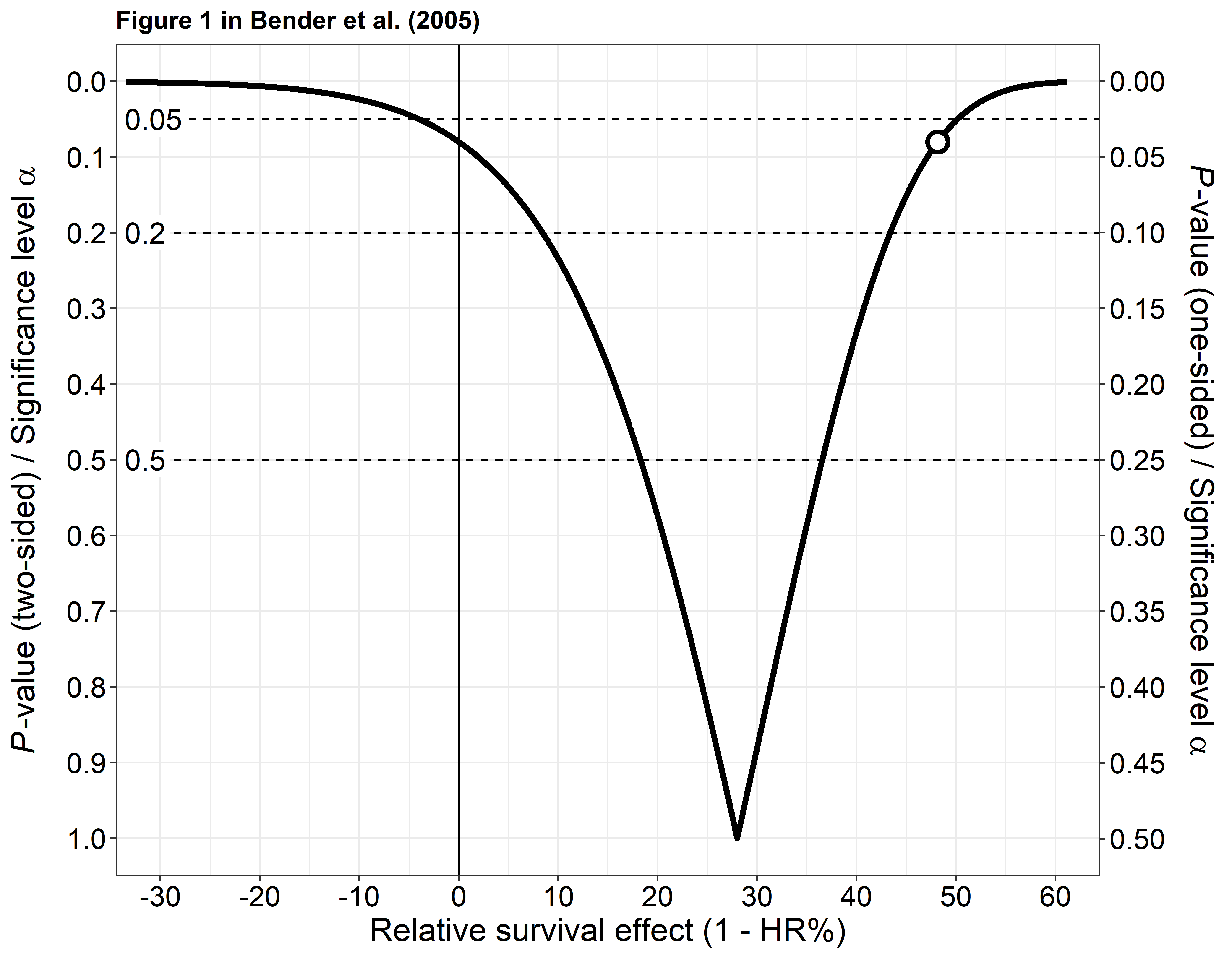
References
Bender R, Berg G, Zeeb H. (2005): Tutorial: using confidence curves in medical research. Biom J. 47(2): 237-47.
Berrar D (2017): Confidence Curves: an alternative to null hypothesis significance testing for the comparison of classifiers. Mach Learn. 106:911-949.
Fraser D. A. S. (2019): The p-value function and statistical inference. Am Stat. 73:sup1, 135-147.
Greenland S (2019): Valid P-Values Behave Exactly as They Should: Some Misleading Criticisms of P-Values and Their Resolution with S-Values. Am Stat. 73sup1, 106-114.
Infanger D, Schmidt-Trucksäss A. (2019): P value functions: An underused method to present research results and to promote quantitative reasoning. Stat Med. 38, 4189-4197. doi: 10.1002/sim.8293.
Poole C. (1987a): Beyond the confidence interval. Am J Public Health. 77(2): 195-9.
Poole C. (1987b): Confidence intervals exclude nothing. Am J Public Health. 77(4): 492-3.
Rafi Z, Greenland S. (2020): Semantic and cognitive tools to aid statistical science: replace confidence and significance by compatibility and surprise. BMC Med Res Methodol. 20, 244. doi: 10.1186/s12874-020-01105-9.
Rosenthal R, Rubin DB. (1994): The counternull value of an effect size: A new statistic. Psychol Sci. 5(6): 329-34.
Schweder T, Hjort NL. (2016): Confidence, likelihood, probability: statistical inference with confidence distributions. New York, NY: Cambridge University Press.
Xie M, Singh K, Strawderman WE. (2011): Confidence Distributions and a Unifying Framework for Meta-Analysis. J Am Stat Assoc. 106(493): 320-33. doi: 10.1198/jasa.2011.tm09803.
Xie Mg, Singh K. (2013): Confidence distribution, the frequentist distribution estimator of a parameter: A review. Internat Statist Rev. 81(1): 3-39.
Contact
Session info
#> R version 4.0.5 (2021-03-31)
#> Platform: x86_64-w64-mingw32/x64 (64-bit)
#> Running under: Windows 10 x64 (build 19042)
#>
#> Matrix products: default
#>
#> locale:
#> [1] LC_COLLATE=German_Switzerland.1252 LC_CTYPE=German_Switzerland.1252
#> [3] LC_MONETARY=German_Switzerland.1252 LC_NUMERIC=C
#> [5] LC_TIME=German_Switzerland.1252
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] pvaluefunctions_1.6.2
#>
#> loaded via a namespace (and not attached):
#> [1] pracma_2.3.3 pillar_1.6.0 compiler_4.0.5 highr_0.9
#> [5] tools_4.0.5 digest_0.6.27 evaluate_0.14 lifecycle_1.0.0
#> [9] tibble_3.1.1 gtable_0.3.0 pkgconfig_2.0.3 rlang_0.4.11
#> [13] DBI_1.1.1 yaml_2.2.1 xfun_0.23 stringr_1.4.0
#> [17] dplyr_1.0.6 knitr_1.33 generics_0.1.0 vctrs_0.3.8
#> [21] grid_4.0.5 tidyselect_1.1.1 glue_1.4.2 R6_2.5.0
#> [25] fansi_0.4.2 rmarkdown_2.8 ggplot2_3.3.3 purrr_0.3.4
#> [29] farver_2.1.0 magrittr_2.0.1 scales_1.1.1 ellipsis_0.3.2
#> [33] htmltools_0.5.1.1 assertthat_0.2.1 colorspace_2.0-1 utf8_1.2.1
#> [37] stringi_1.6.1 munsell_0.5.0 crayon_1.4.1
