Description
Quantile G-Computation.
Description
G-computation for a set of time-fixed exposures with quantile-based basis functions, possibly under linearity and homogeneity assumptions. This approach estimates a regression line corresponding to the expected change in the outcome (on the link basis) given a simultaneous increase in the quantile-based category for all exposures. Works with continuous, binary, and right-censored time-to-event outcomes. Reference: Alexander P. Keil, Jessie P. Buckley, Katie M. OBrien, Kelly K. Ferguson, Shanshan Zhao, and Alexandra J. White (2019) A quantile-based g-computation approach to addressing the effects of exposure mixtures; <doi:10.1289/EHP5838>.
README.md
qgcomp v2.15.2
QGcomp (quantile g-computation): estimating the effects of exposure mixtures. Works for continuous, binary, and right-censored survival outcomes.
Flexible, unconstrained, fast and guided by modern causal inference principles
Quick start
# install developers version (requires devtools)
# install.packages("devtools")
# devtools::install_github("alexpkeil1/qgcomp") # master version (usually reliable)
# devtools::install_github("alexpkeil1/qgcomp@dev") # dev version (may not be working)
# or install version from CRAN
install.packages("qgcomp")
library("qgcomp")
# using data from the qgcomp package
data("metals", package="qgcomp")
Xnm <- c(
'arsenic','barium','cadmium','calcium','chromium','copper',
'iron','lead','magnesium','manganese','mercury','selenium','silver',
'sodium','zinc'
)
# continuous outcome
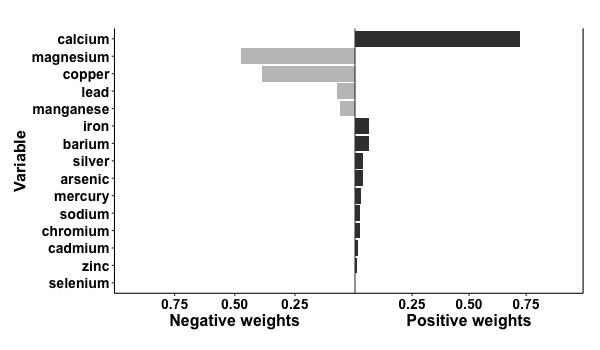
results = qgcomp.noboot(y~.,dat=metals[,c(Xnm, 'y')], family=gaussian())
print(results)
> Scaled effect size (positive direction, sum of positive coefficients = 0.39)
> calcium iron barium silver arsenic mercury sodium chromium cadmium zinc
> 0.72216 0.06187 0.05947 0.03508 0.03447 0.02451 0.02162 0.02057 0.01328 0.00696
>
> Scaled effect size (negative direction, sum of negative coefficients = -0.124)
> magnesium copper lead manganese selenium
> 0.475999 0.385299 0.074019 0.063828 0.000857
>
> Mixture slope parameters (Delta method CI):
>
> Estimate Std. Error Lower CI Upper CI t value Pr(>|t|)
> (Intercept) -0.356670 0.107878 -0.56811 -0.14523 -3.3062 0.0010238
> psi1 0.266394 0.071025 0.12719 0.40560 3.7507 0.0002001
p = plot(results, suppressprint=TRUE)
ggplot2::ggsave("res1.png", plot=p, dpi=72, width=600/72, height=350/72, units="in")
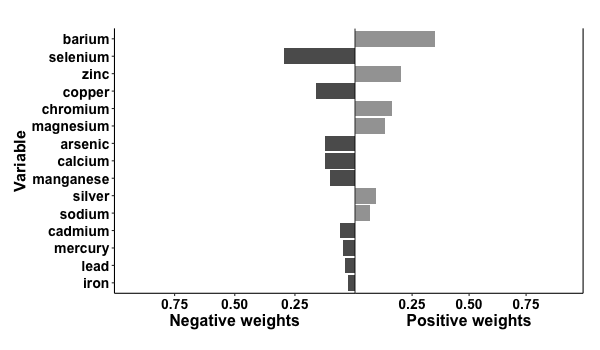
# binary outcome
results2 = qgcomp.noboot(disease_state~., expnms=Xnm,
data = metals[,c(Xnm, 'disease_state')], family=binomial(), q=4)
print(results2)
> Scaled effect size (positive direction, sum of positive coefficients = 0.392)
> barium zinc chromium magnesium silver sodium
> 0.3520 0.2002 0.1603 0.1292 0.0937 0.0645
>
> Scaled effect size (negative direction, sum of negative coefficients = -0.696)
> selenium copper arsenic calcium manganese cadmium mercury lead iron
> 0.2969 0.1627 0.1272 0.1233 0.1033 0.0643 0.0485 0.0430 0.0309
>
> Mixture log(OR) (Delta method CI):
>
> Estimate Std. Error Lower CI Upper CI Z value Pr(>|z|)
> (Intercept) 0.26362 0.51615 -0.74802 1.27526 0.5107 0.6095
> psi1 -0.30416 0.34018 -0.97090 0.36258 -0.8941 0.3713
p2 = plot(results2, suppressprint=TRUE)
ggplot2::ggsave("res2.png", plot=p2, dpi=72, width=600/72, height=350/72, units="in")
adjusting for covariate(s)
results3 = qgcomp.noboot(y ~ mage35 + arsenic + barium + cadmium + calcium + chloride +
chromium + copper + iron + lead + magnesium + manganese +
mercury + selenium + silver + sodium + zinc,
expnms=Xnm,
metals, family=gaussian(), q=4)
print(results3)
> Scaled effect size (positive direction, sum of positive coefficients = 0.381)
> calcium barium iron silver arsenic mercury chromium zinc sodium cadmium
> 0.74466 0.06636 0.04839 0.03765 0.02823 0.02705 0.02344 0.01103 0.00775 0.00543
>
> Scaled effect size (negative direction, sum of negative coefficients = -0.124)
> magnesium copper lead manganese selenium
> 0.49578 0.35446 0.08511 0.06094 0.00372
>
> Mixture slope parameters (Delta method CI):
>
> Estimate Std. Error Lower CI Upper CI t value Pr(>|t|)
> (Intercept) -0.348084 0.108037 -0.55983 -0.13634 -3.2219 0.0013688
> psi1 0.256969 0.071459 0.11691 0.39703 3.5960 0.0003601
# coefficient for confounder
results3$fit$coefficients['mage35']
> mage35
> 0.03463519
Bootstrapping to get population average risk ratio via g-computation using qgcomp.boot
results4 = qgcomp.boot(disease_state~., expnms=Xnm,
data = metals[,c(Xnm, 'disease_state')], family=binomial(),
q=4, B=1000,# B should be 200-500+ in practice
seed=125, rr=TRUE)
print(results4)
> Mixture log(RR) (bootstrap CI):
>
> Estimate Std. Error Lower CI Upper CI Z value Pr(>|z|)
> (Intercept) -0.56237 0.23773 -1.02832 -0.096421 -2.3655 0.0180
> psi1 -0.16373 0.17239 -0.50161 0.174158 -0.9497 0.3423
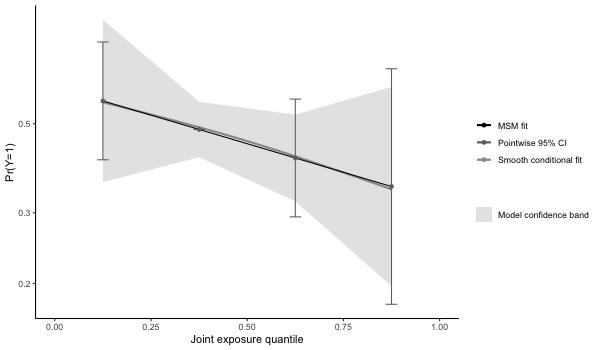
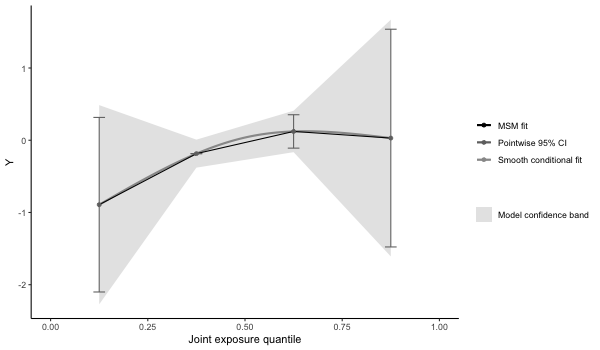
# checking whether model fit seems appropriate (note that the conditional fit
# appears slightly non-linear. The conditional model is on the log-odds scale
# wheras the marginal structural model is on the log-probability scale ).
# The plot is on the log-10 scale.
p4 = plot(results4, suppressprint=TRUE)
ggplot2::ggsave("res4.png", plot=p4, dpi=72, width=600/72, height=350/72, units="in")
Allowing for interactions and non-linear terms using qgcomp.boot
results5 = qgcomp(y~. + .^2 + arsenic*cadmium,
expnms=Xnm,
metals[,c(Xnm, 'y')], family=gaussian(), q=4, B=10,
seed=125, degree=2)
print(results5)
> Mixture slope parameters (bootstrap CI):
>
> Estimate Std. Error Lower CI Upper CI t value Pr(>|t|)
> (Intercept) -0.89239 0.70336 -2.27095 0.48617 -1.2688 0.2055
> psi1 0.90649 0.93820 -0.93235 2.74533 0.9662 0.3347
> psi2 -0.19970 0.32507 -0.83682 0.43743 -0.6143 0.5395
# some apparent non-linearity, but would require more bootstrap iterations for
# proper test of non-linear mixture effect
p5 = plot(results5, suppressprint=TRUE)
ggplot2::ggsave("res5.png", plot=p5, dpi=72, width=600/72, height=350/72, units="in")
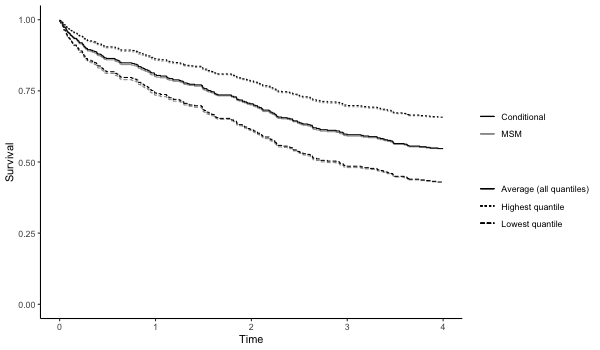
Survival outcomes with and without bootstrapping (fitting a marginal structural cox model to estimate the hazard ratio)
results6 = qgcomp.cox.noboot(Surv(disease_time, disease_state)~.,
expnms=Xnm,
metals[,c(Xnm, 'disease_time', 'disease_state')])
print(results6)
> Scaled effect size (positive direction, sum of positive coefficients = 0.32)
> barium zinc magnesium chromium silver sodium iron
> 0.3432 0.1946 0.1917 0.1119 0.0924 0.0511 0.0151
>
> Scaled effect size (negative direction, sum of negative coefficients = -0.554)
> selenium copper calcium arsenic manganese cadmium lead mercury
> 0.2705 0.1826 0.1666 0.1085 0.0974 0.0794 0.0483 0.0466
>
> Mixture log(hazard ratio) (Delta method CI):
>
> Estimate Std. Error Lower CI Upper CI Z value Pr(>|z|)
> psi1 -0.23356 0.24535 -0.71444 0.24732 -0.9519 0.3411
results7 = qgcomp.cox.boot(Surv(disease_time, disease_state)~.,
expnms=Xnm,
metals[,c(Xnm, 'disease_time', 'disease_state')],
B=10, MCsize=5000)
p7 = plot(results7, suppressprint=TRUE)
ggplot2::ggsave("res7.png", plot=p7, dpi=72, width=600/72, height=350/72, units="in")
More help
See the vignette which is included with the qgcomp R package, and is accessible in R via vignette("qgcomp-vignette", package="qgcomp")