Estimation of Natural Regeneration Potential.
quaxnat 
An R package for estimating the regeneration potential using large scale inventory information.
Description
Regeneration datasets, especially those pertaining to rare tree species, often exhibit values that are below the real regeneration potential due to various factors influencing or hindering natural regeneration. The R package quaxnat implements a quantile regression approach for estimating the regeneration potential, following the methodology of Axer et al. (2021). This quantile regression method quantifies the impact of the distance to the nearest seed source on natural regeneration densities not considering other influencing variables. The function quax we provide is a simple implementation of the quantile estimation. It presents a rather naive approach to quantile regression and, while likely outperformed by more sophisticated procedures such as those in the R package quantreg(Koenker et al., 2023) in general quantile regression problems, appears to work reasonably well in the situation considered here.
The package provides various dispersal kernels that have proven useful in modeling seed dispersal:
- Spatial t distribution
- Weibull distribution
- Lognormal distribution
- Power distribution
- Exponential power distribution
Within the quax function the dispersal kernel is defined by fun. Additional kernels can be provided as user-defined functions.
Installation
The stable version can be installed from CRAN
install.packages("quaxnat")
and the development version is available from Github
remotes::install_github("MaximilianAxer/quaxnat")
Usage
This section illustrates the use of the quax function with step-by-step explanations of the computation.
# Load quaxnat package
library(quaxnat)
# Load example data
data("regeneration")
After loading the quaxnat package, we are now ready to apply the quax function, which we do for example for the .995th quantile and five dispersal kernels implemented in quaxnat.
# Estimate regeneration potential based on various dispersal kernels
tau <- 0.995
f1 <- quax(oak_regen ~ distance_oak, regeneration, tau=tau, fun=k_t)
f2 <- quax(oak_regen ~ distance_oak, regeneration, tau=tau, fun=k_weibull)
f3 <- quax(oak_regen ~ distance_oak, regeneration, tau=tau, fun=k_lognormal)
f4 <- quax(oak_regen ~ distance_oak, regeneration, tau=tau, fun=k_power)
f5 <- quax(oak_regen ~ distance_oak, regeneration, tau=tau, fun=k_exponential_power)
f <- list(`Spatial t`=f1, Weibull=f2, Lognormal=f3, Power=f4, `Exp. Power`=f5)
The results of the quantile regression for the different dispersal kernels can be visualised and compared. We are now ready to add the estimated functions to the plot. Since quax directly returns those functions, this is conveniently done with the curve function.
# Plot regeneration density as a function of the distance to the nearest seed tree
plot(oak_regen ~ distance_oak, regeneration, xlim=c(0, 1500), cex=0.8,
xlab="Distance to nearest seed tree", ylab="Regeneration density")
# Add estimated functions to diagram
col <- hcl.colors(length(f), palette="Dynamic")
for (i in seq_along(f)){
curve(f[[i]](x), add=TRUE, n=10000, col=col[i], lwd=2)
}
legend("topright", title=paste0(tau," quantile"),
legend=names(f), lty=1, lwd=2, col=col)
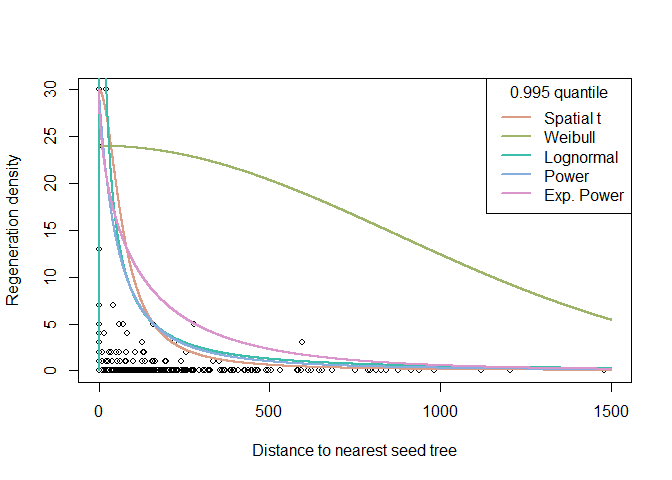
For the quax objects returned by the quax function, summary outputs the estimated coefficients and the attained value of the objective function. The latter can be used to compare fits of different dispersal kernels for the same quantile to the same data.
# Compare quality of fits
sapply(f, summary)
#> Spatial t Weibull Lognormal Power Exp. Power
#> coefficients numeric,2 numeric,2 numeric,2 numeric,2 numeric,2
#> value 45.99978 151.7906 131.241 45.92098 46.46311
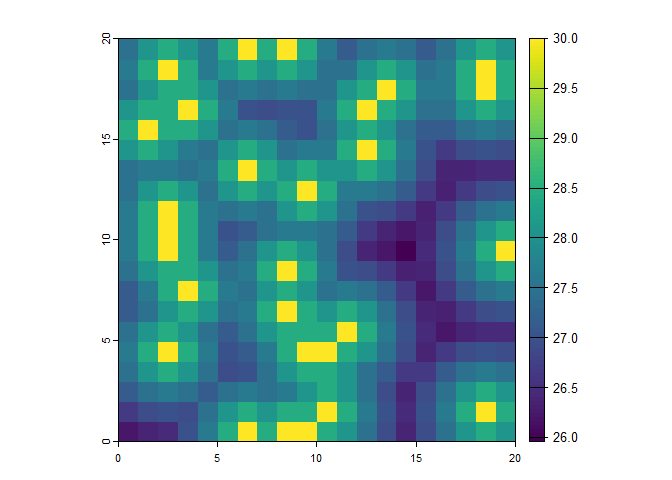
A prediction for the entire study area is made with the predict_quax function. A distmap with distances to the nearest seed tree is used for the prediction of the potential regeneration densities. Furthermore, the parameterized quax object is used for the prediction.
# Create raster data set as prediction area
set.seed(20240606)
rr <- terra::rast(matrix(sample(0:10, 20 * 20, replace = TRUE), nrow = 20, ncol = 20))
# Compute distances to seed trees for prediction area
dists <- seed_tree_distmap(raster = rr, species = "10")
# Predict the potential regeneration density
regen_dens <- predict_quax(dists, f5)
terra::plot(regen_dens)
Authors and contributors
- Maximilian Axer (main author of the package)
- Robert Schlicht (author of the package)
- Robert Nuske (help with package development)
License
GPL (>=2)
Literature
Axer et al. “Modelling potential density of natural regeneration of European oak species (Quercus robur L., Quercus petraea (Matt.) Liebl.) depending on the distance to the potential seed source: Methodological approach for modelling dispersal from inventory data at forest enterprise level.” Forest Ecology and Management 482 (2021): 118802.
Koenker et al. “Package ‘quantreg’.” Reference manual available at CRAN: https://cran.r-project.org/package=quantreg (2023).