REST API Client for the 'PGS' Catalog.
quincunx 
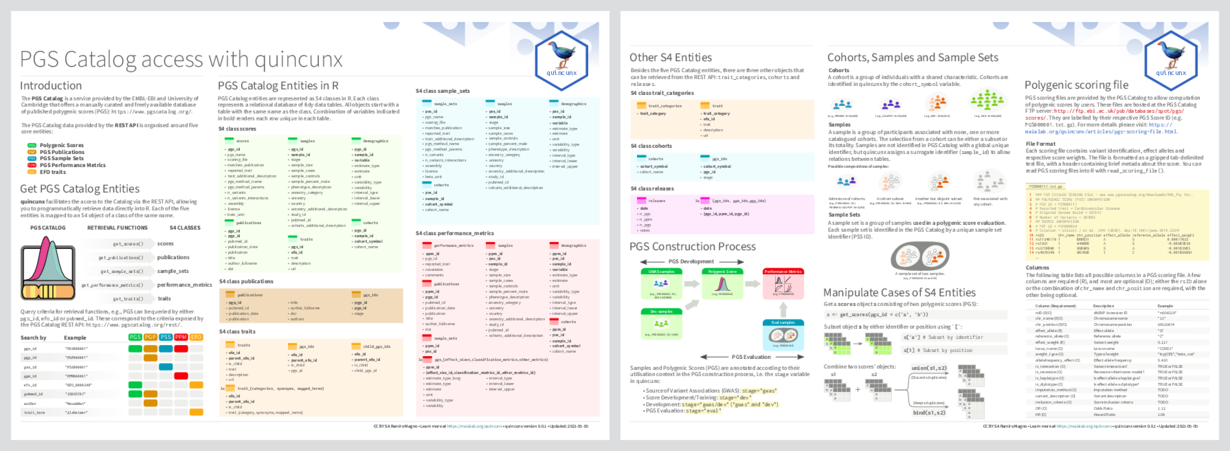
The goal of {quincunx} is to provide programmatic access to the Polygenic Score (PGS) Catalog, an open resource for polygenic scores and associated metadata describing their development and respective assessment.
Browse the online documentation at rmagno.eu/quincunx to get started.
Installation
Install {quincunx} from CRAN:
# Install from CRAN
install.packages("quincunx")
You can instead install the development version with:
# install.packages("pak")
pak::pak("ramiromagno/quincunx")
Cheatsheet
Citing this work
{quincunx} was published in Bioinformatics in 2021: https://doi.org/10.1093/bioinformatics/btab522.
To generate a citation for this publication from within R:
citation('quincunx')
#> To cite quincunx in publications use:
#>
#> Ramiro Magno, Isabel Duarte, Ana-Teresa Maia, quincunx: an R package
#> to query, download and wrangle PGS Catalog data, Bioinformatics,
#> btab522, 16 July 2021, Pages 1-3,
#> https://doi.org/10.1093/bioinformatics/btab522
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Article{,
#> title = {quincunx: an R package to query, download and wrangle PGS Catalog data},
#> author = {Ramiro Magno and Isabel Duarte and Ana-Teresa Maia},
#> journal = {Bioinformatics},
#> year = {2021},
#> volume = {38},
#> number = {1},
#> pages = {1--3},
#> url = {https://doi.org/10.1093/bioinformatics/btab522},
#> }
Citing PGS Catalog publications
Also, please do not forget to cite the authors behind the original studies and the papers associated with the PGS Catalog project:
- Lambert, S.A., Gil, L., Jupp, S. et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat Genet 53, 420–425 (2021). doi: 10.1038/s41588-021-00783-5
- Wand, H., Lambert, S.A., Tamburro, C. et al. Improving reporting standards for polygenic scores in risk prediction studies. Nature 591, 211–219 (2021). doi: 10.1038/s41586-021-03243-6
Terms of use
Please note that if you use the data provided by the PGS Catalog either directly or via {quincunx} you agree to abide to the EMBL-EBI Terms of Use.
Code of Conduct
Please note that the {quincunx} project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
Acknowledgements
This work would have not been possible without the precious feedback from the PGS Catalog team, particularly Samuel Lambert and Laurent Gil.
Package name and hex sticker
The package name {quincunx} is another name for Galton Board, that so nicely exemplifies the Central Limit Theorem, which in turn is a key concept of genetics, namely, Fisher’s infinitesimal model… which leads us to Polygenic Scores, the key concept of the PGS Catalog.
The bird in the hex sticker is a Porphyrio porphyrio, an emblematic species native to the Ria Formosa Natural Park, which is a wildlife reserve surrounding the University of Algarve, where the authors are affiliated.