Description
Synchrony in Motion Energy Analysis (MEA) Time-Series.
Description
A suite of tools useful to read, visualize and export bivariate motion energy time-series. Lagged synchrony between subjects can be analyzed through windowed cross-correlation. Surrogate data generation allows an estimation of pseudosynchrony that helps to estimate the effect size of the observed synchronization. Kleinbub, J. R., & Ramseyer, F. T. (2020). rMEA: An R package to assess nonverbal synchronization in motion energy analysis time-series. Psychotherapy research, 1-14. <doi:10.1080/10503307.2020.1844334>.
README.md
rMEA
The goal of rMEA is to provide a suite of tools useful to read, visualize and export bivariate Motion Energy time-series. Lagged synchrony between subjects can be analyzed through windowed cross-correlation. Surrogate data generation allows an estimation of pseudosynchrony that helps to estimate the effect size of the observed synchronization.
Example
This example shows a complete analysis pipeline consisting on Motion Energy time-series import, pre-processing, cross-correlation analysis and comparison between groups against pseudosynchrony.
library(rMEA)
## read the first sample (intake interviews of patients that carried on therapy)
path_normal <- system.file("extdata/normal", package = "rMEA")
mea_normal <- readMEA(path_normal, sampRate = 25, s1Col = 1, s2Col = 2,
s1Name = "Patient", s2Name = "Therapist", skip=1,
idOrder = c("id","session"), idSep="_")
#>
#> STEP 1 | Reading 10 dyads
#> ..................................................|100%
#> ..................................................|Done ;)
#>
#> STEP 2 | Formatting data frames:
#> ..................................................|100%
#> ..................................................|Done ;)
#> Warning: 0.07% of the data was higher than 10 standard deviations in dyad:
#> 200, session: 1, group:all. Check the raw data!
#> Warning: 0.03% of the data was higher than 10 standard deviations in dyad:
#> 201, session: 1, group:all. Check the raw data!
#> Warning: 0.03% of the data was higher than 10 standard deviations in dyad:
#> 202, session: 1, group:all. Check the raw data!
#> Warning: 0.04% of the data was higher than 10 standard deviations in dyad:
#> 204, session: 1, group:all. Check the raw data!
#> Warning: 0.01% of the data was higher than 10 standard deviations in dyad:
#> 205, session: 1, group:all. Check the raw data!
#> Warning: 0.05% of the data was higher than 10 standard deviations in dyad:
#> 206, session: 1, group:all. Check the raw data!
#> Warning: 0.01% of the data was higher than 10 standard deviations in dyad:
#> 208, session: 1, group:all. Check the raw data!
#>
#> STEP 3 | ReadMEA report
#> Filename id_dyad session group duration_hh.mm.ss Patient_%
#> 1 200_01.txt 200 1 all 00:10:00 50.1
#> 2 201_01.txt 201 1 all 00:10:00 50.2
#> 3 202_01.txt 202 1 all 00:10:00 44.6
#> 4 203_01.txt 203 1 all 00:10:00 97.4
#> 5 204_01.txt 204 1 all 00:10:00 66.4
#> 6 205_01.txt 205 1 all 00:10:00 84.3
#> 7 206_01.txt 206 1 all 00:10:00 30.3
#> 8 207_01.txt 207 1 all 00:10:00 52.5
#> 9 208_01.txt 208 1 all 00:10:00 76.6
#> 10 209_01.txt 209 1 all 00:10:00 72.4
#> Therapist_%
#> 1 59.5
#> 2 74.7
#> 3 47.1
#> 4 73.1
#> 5 55.7
#> 6 69.9
#> 7 34.4
#> 8 42.3
#> 9 44.2
#> 10 63.1
mea_normal <- setGroup(mea_normal, "normal")
## read the second sample (intake interviews of patients that dropped out)
path_dropout <- system.file("extdata/dropout", package = "rMEA")
mea_dropout <- readMEA(path_dropout, sampRate = 25, s1Col = 1, s2Col = 2,
s1Name = "Patient", s2Name = "Therapist", skip=1,
idOrder = c("id","session"), idSep="_")
#>
#> STEP 1 | Reading 10 dyads
#> ..................................................|100%
#> ..................................................|Done ;)
#>
#> STEP 2 | Formatting data frames:
#> ..................................................|100%
#> ..................................................|Done ;)
#> Warning: 0.03% of the data was higher than 10 standard deviations in dyad:
#> 100, session: 1, group:all. Check the raw data!
#> Warning: 0.01% of the data was higher than 10 standard deviations in dyad:
#> 101, session: 1, group:all. Check the raw data!
#> Warning: 0.09% of the data was higher than 10 standard deviations in dyad:
#> 104, session: 1, group:all. Check the raw data!
#> Warning: 0.02% of the data was higher than 10 standard deviations in dyad:
#> 105, session: 1, group:all. Check the raw data!
#> Warning: 0.1% of the data was higher than 10 standard deviations in dyad:
#> 106, session: 1, group:all. Check the raw data!
#>
#> STEP 3 | ReadMEA report
#> Filename id_dyad session group duration_hh.mm.ss Patient_%
#> 1 100_01.txt 100 1 all 00:10:00 85.0
#> 2 101_01.txt 101 1 all 00:10:00 84.2
#> 3 102_01.txt 102 1 all 00:10:00 90.9
#> 4 103_01.txt 103 1 all 00:10:00 72.5
#> 5 104_01.txt 104 1 all 00:10:00 72.5
#> 6 105_01.txt 105 1 all 00:10:00 69.2
#> 7 106_01.txt 106 1 all 00:10:00 76.7
#> 8 107_01.txt 107 1 all 00:10:00 95.4
#> 9 108_01.txt 108 1 all 00:10:00 80.1
#> 10 109_01.txt 109 1 all 00:10:00 85.0
#> Therapist_%
#> 1 97.2
#> 2 88.0
#> 3 95.1
#> 4 90.7
#> 5 73.9
#> 6 83.5
#> 7 66.1
#> 8 79.5
#> 9 96.5
#> 10 87.3
mea_dropout <- setGroup(mea_dropout, "dropout")
## Combine into a single object
mea_all <- c(mea_normal, mea_dropout)
summary(mea_all)
#>
#> MEA analysis results:
#> dyad session group Patient_% Therapist_%
#> normal_200_1 200 1 normal 50.1 59.5
#> normal_201_1 201 1 normal 50.2 74.7
#> normal_202_1 202 1 normal 44.6 47.1
#> normal_203_1 203 1 normal 97.4 73.1
#> normal_204_1 204 1 normal 66.4 55.7
#> normal_205_1 205 1 normal 84.3 69.9
#> normal_206_1 206 1 normal 30.3 34.4
#> normal_207_1 207 1 normal 52.5 42.3
#> normal_208_1 208 1 normal 76.6 44.2
#> normal_209_1 209 1 normal 72.4 63.1
#> dropout_100_1 100 1 dropout 85.0 97.2
#> dropout_101_1 101 1 dropout 84.2 88.0
#> dropout_102_1 102 1 dropout 90.9 95.1
#> dropout_103_1 103 1 dropout 72.5 90.7
#> dropout_104_1 104 1 dropout 72.5 73.9
#> dropout_105_1 105 1 dropout 69.2 83.5
#> dropout_106_1 106 1 dropout 76.7 66.1
#> dropout_107_1 107 1 dropout 95.4 79.5
#> dropout_108_1 108 1 dropout 80.1 96.5
#> dropout_109_1 109 1 dropout 85.0 87.3
#>
#> Data processing: raw
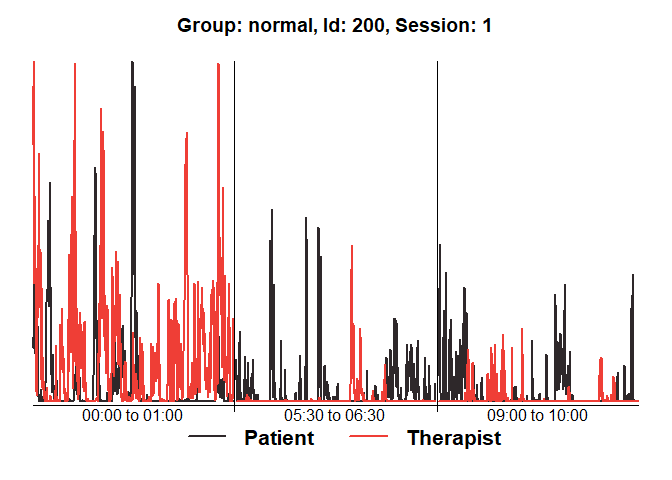
## Show diagnostics for the first session:
diagnosticPlot(mea_all[[1]])
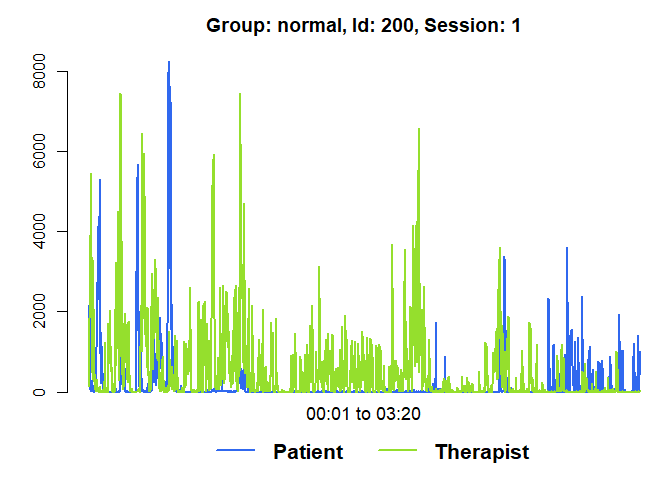
plot(mea_all[[1]], from=1, to=200)
## Filter the data
mea_smoothed <- MEAsmooth(mea_all)
#>
#> Moving average smoothing:
#> ..................................................|100%
#> ..................................................|Done ;)
mea_rescaled <- MEAscale(mea_smoothed)
#>
#> Rescaling data:
#> ..................................................|100%
#> ..................................................|Done ;)
## Generate a random sample
mea_random <- shuffle(mea_rescaled, 50)
#>
#> Shuffling dyads:
#> ..................................................|100%
#> ..................................................|Done ;)
#>
#> 50 / 760 possible combinations were randomly selected
## Run CCF analysis
mea_ccf <- MEAccf(mea_rescaled, lagSec= 5, winSec = 30, incSec=10, ABS = F)
#>
#> Computing CCF:
#> ..................................................|100%
#> ..................................................|Done ;)
mea_random_ccf <- MEAccf(mea_random, lagSec= 5, winSec = 30, incSec=10, ABS = F)
#>
#> Computing CCF:
#> ..................................................|100%
#> ..................................................|Done ;)
## Visualize results
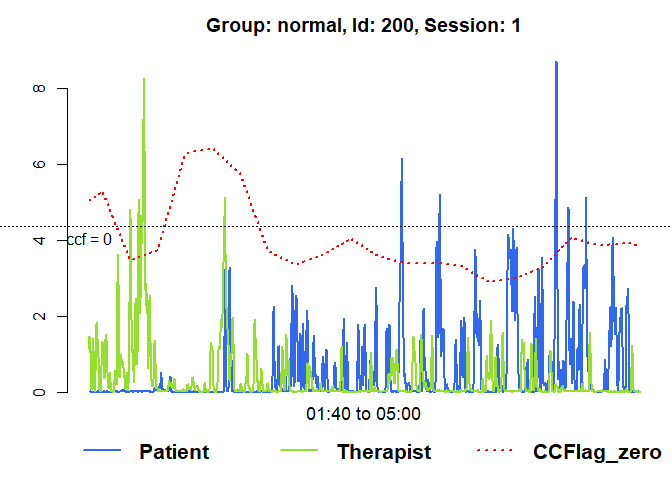
# Raw data of the first session with running lag-0 ccf
plot(mea_ccf[[1]], from=100, to=300, ccf = "lag_zero")
# Heatmap of the first session
MEAheatmap(mea_ccf[[1]])
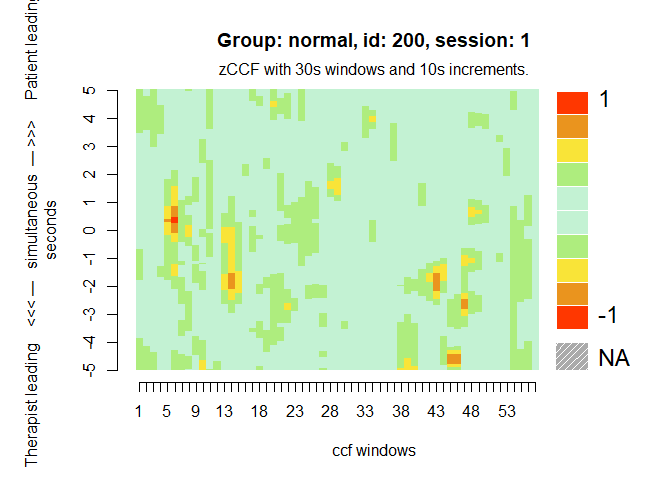
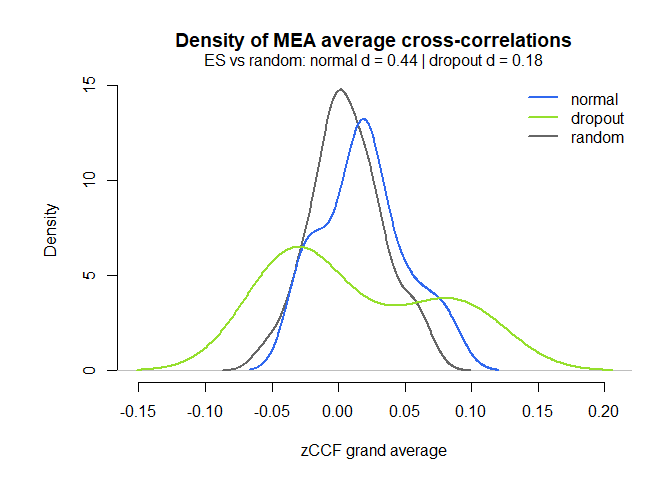
# Distribution of the ccf calculations by group, against random matched dyads
MEAdistplot(mea_ccf, contrast = mea_random_ccf)
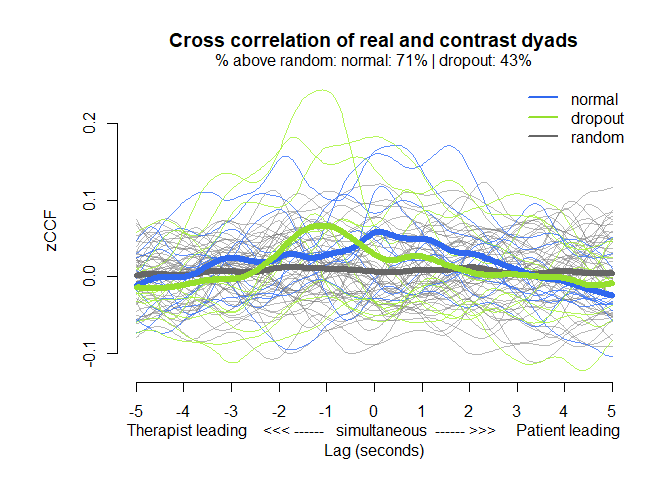
# Representation of the average cross-correlations by lag
MEAlagplot(mea_ccf, contrast=mea_random_ccf)