Description
Spectral Modularity Clustering.
Description
Implements the network clustering algorithm described in Newman (2006) <doi:10.1103/PhysRevE.74.036104>. The complete iterative algorithm comprises of two steps. In the first step, the network is expressed in terms of its leading eigenvalue and eigenvector and recursively partition into two communities. Partitioning occurs if the maximum positive eigenvalue is greater than the tolerance (10e-5) for the current partition, and if it results in a positive contribution to the Modularity. Given an initial separation using the leading eigen step, 'rSpectral' then continues to maximise for the change in Modularity using a fine-tuning step - or variate thereof. The first stage here is to find the node which, when moved from one community to another, gives the maximum change in Modularity. This node’s community is then fixed and we repeat the process until all nodes have been moved. The whole process is repeated from this new state until the change in the Modularity, between the new and old state, is less than the predefined tolerance. A slight variant of the fine-tuning step, which can improve speed of the calculation, is also provided. Instead of moving each node into each community in turn, we only consider moves of neighbouring nodes, found in different communities, to the community of the current node of interest. The two steps process is repeatedly applied to each new community found, subdivided each community into two new communities, until we are unable to find any division that results in a positive change in Modularity.
README.md
rSpectral
The goal of ‘rSpectral’ is to make Spectral Modularity graph clustering method available to most of R graph frameworks.
Installation
You can install the development version of rSpectral from GitHub with:
# install.packages("devtools")
devtools::install_github("cmclean5/rSpectral")
Example
This is a basic example which shows you how to solve a common problem
- load the karate club graph and plot Faction membership:
library(rSpectral)
library(igraph)
#>
#> Attaching package: 'igraph'
#> The following objects are masked from 'package:stats':
#>
#> decompose, spectrum
#> The following object is masked from 'package:base':
#>
#> union
data(karate, package="igraphdata")
l<-layout_nicely(karate)
memT<-V(karate)$Faction
palette <- rainbow(max(as.numeric(memT)))
plot(karate,vertex.color=palette[memT],layout=l)
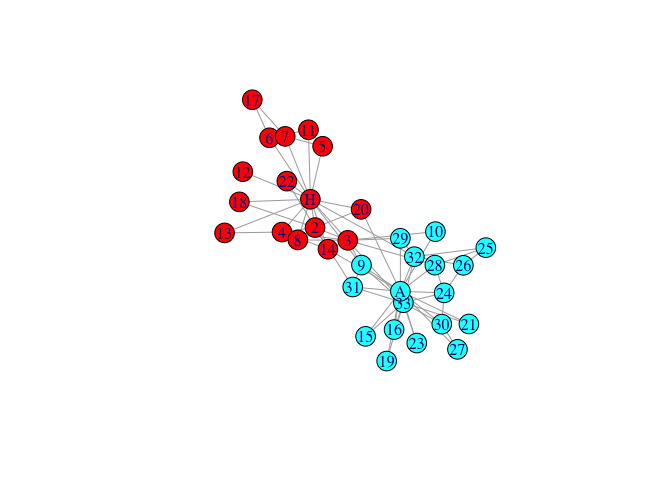
- run spectral clustering on graph
mem0<-igraph::membership(rSpectral::spectral_igraph_communities(karate))
- plot the graph with membership colors
palette <- rainbow(max(as.numeric(mem0)))
plot(karate,vertex.color=palette[mem0],layout=l)
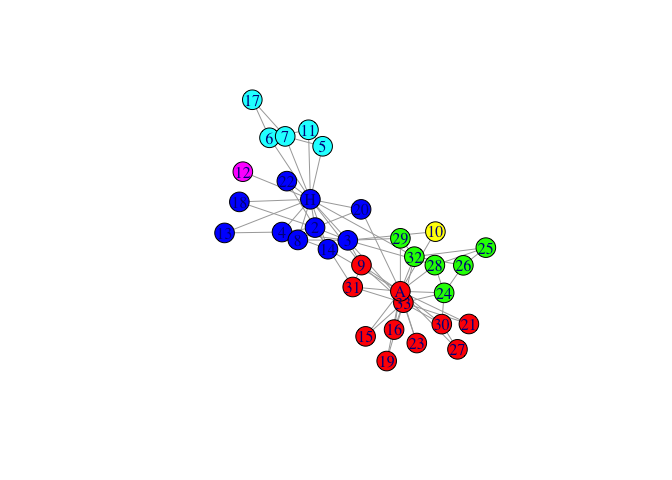
- run spectral clustering on graph, fixing neighbouring nodes found in same community
mem1<-igraph::membership(
rSpectral::spectral_igraph_communities(karate, fix_neig=1))
- and plot again
palette <- rainbow(max(as.numeric(mem1)))
plot(karate,vertex.color=palette[mem1],layout=l)
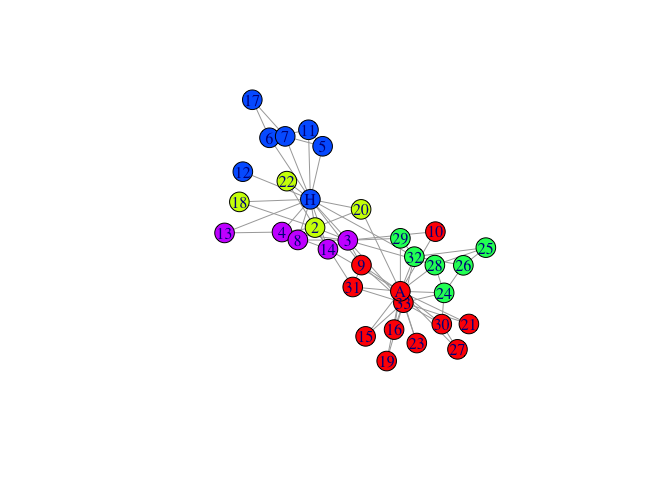
- run spectral clustering - fixing neighbouring nodes, Cnmin=5
mem1.5<-igraph::membership(
rSpectral::spectral_igraph_communities(karate, fix_neig=1,Cn_min=5))
- and the last plot
palette <- rainbow(max(as.numeric(mem1.5)))
plot(karate,vertex.color=palette[mem1.5],layout=l)
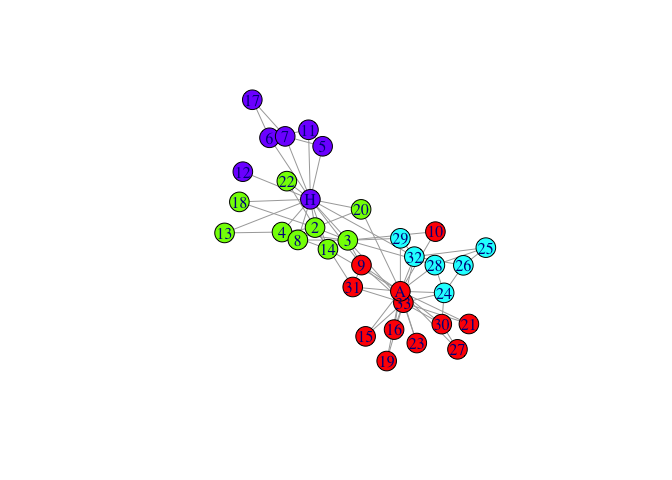
GraphNEL objects could be processed similarily, all other graph types could be converted either to igraph or to GraphNEL by packages such as Intergraph.