The Reinert Method for Textual Data Clustering.
Rainette
Rainette is an R package which implements a variant of the Reinert textual clustering method. This method is available in other softwares such as Iramuteq (free software) or Alceste (commercial, closed source).
Features
- Simple and double clustering algorithms
- Plot functions and shiny interfaces to visualise and explore clustering results
- Utility functions to split a corpus into segments or import a corpus in Iramuteq format
Installation
The package is installable from CRAN.
install_packages("rainette")
The development version is installable from R-universe.
install.packages("rainette", repos = "https://juba.r-universe.dev")
Usage
Let's start with an example corpus provided by the excellent quanteda package.
library(quanteda)
data_corpus_inaugural
First, we'll use split_segments() to split each document into segments of about 40 words (punctuation is taken into account).
corpus <- split_segments(data_corpus_inaugural, segment_size = 40)
Next, we'll apply some preprocessing and compute a document-term matrix with quanteda functions.
tok <- tokens(corpus, remove_punct = TRUE)
tok <- tokens_remove(tok, stopwords("en"))
dtm <- dfm(tok, tolower = TRUE)
dtm <- dfm_trim(dtm, min_docfreq = 10)
We can then apply a simple clustering on this matrix with the rainette() function. We specify the number of clusters (k), and the minimum number of forms in each segment (min_segment_size). Segments which do not include enough forms will be merged with the following or previous one when possible.
res <- rainette(dtm, k = 6, min_segment_size = 15)
We can use the rainette_explor() shiny interface to visualise and explore the different clusterings at each k.
rainette_explor(res, dtm, corpus)
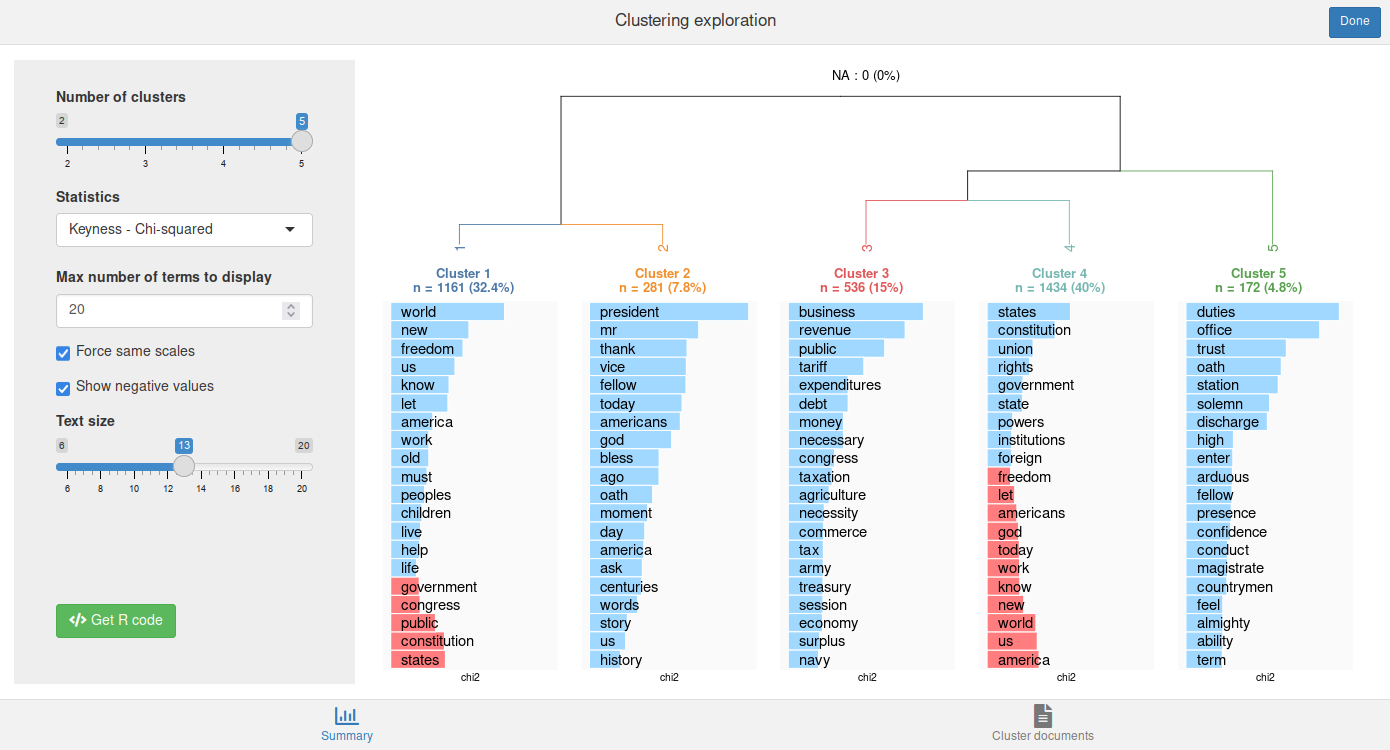
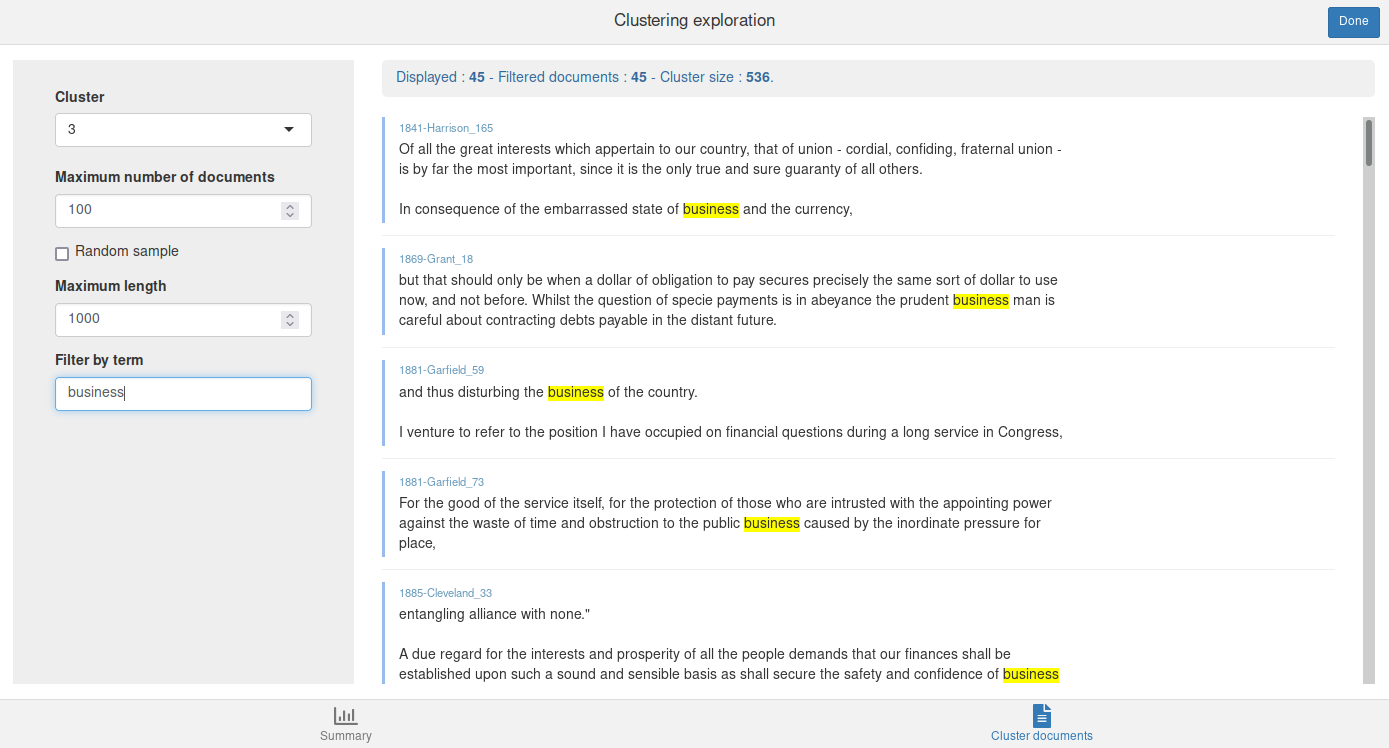
The Cluster documents tab allows to browse and filter the documents in each cluster.
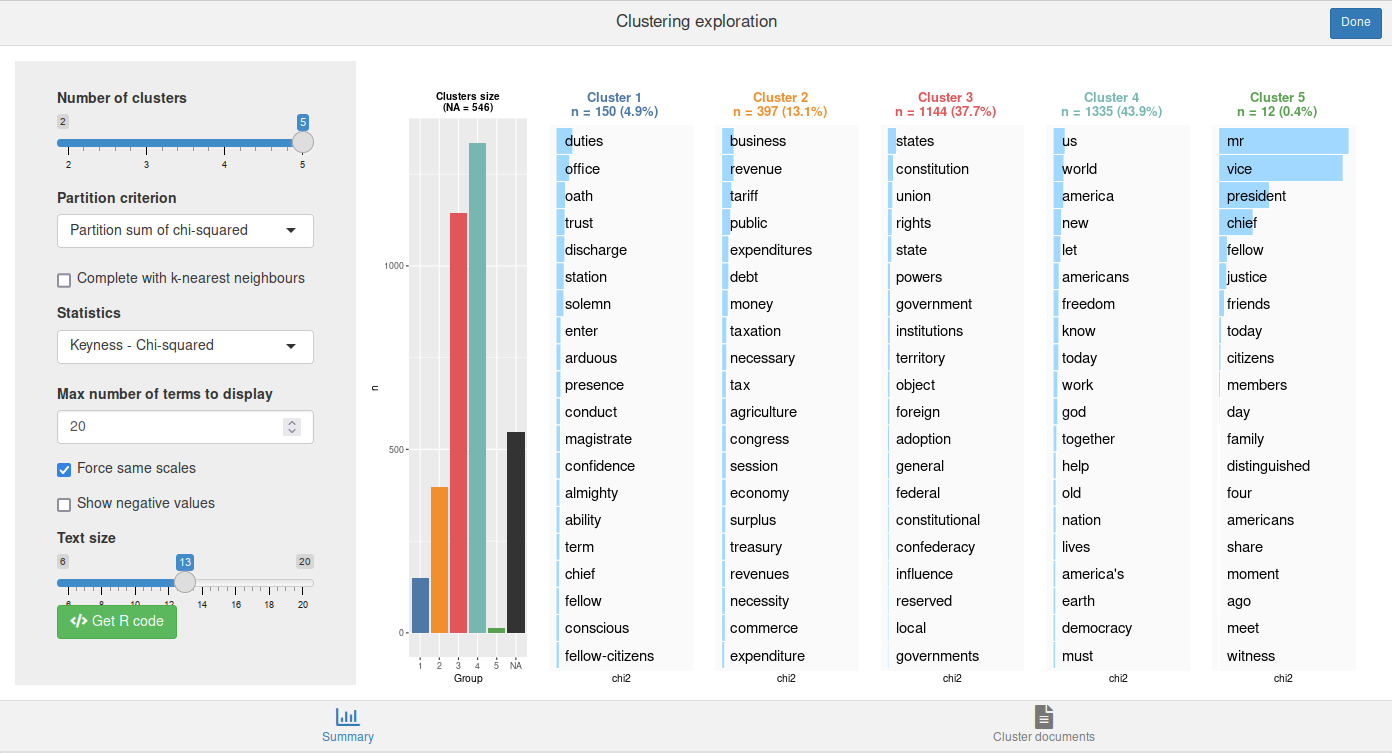
We can also directly generate the clusters description plot for a given k with rainette_plot().
rainette_plot(res, dtm, k = 5)
Or cut the tree at chosen k and add a group membership variable to our corpus metadata.
docvars(corpus)$cluster <- cutree(res, k = 5)
In addition to this, we can also perform a double clustering, ie two simple clusterings produced with different min_segment_size which are then "crossed" to generate more robust clusters. To do this, we use rainette2() on two rainette() results :
res1 <- rainette(dtm, k = 5, min_segment_size = 10)
res2 <- rainette(dtm, k = 5, min_segment_size = 15)
res <- rainette2(res1, res2, max_k = 5)
We can then use rainette2_explor() to explore and visualise the results.
rainette2_explor(res, dtm, corpus)
Tell me more
Two vignettes are available :
Credits
This clustering method has been created by Max Reinert, and is described in several articles, notably :
- Reinert M., "Une méthode de classification descendante hiérarchique : application à l'analyse lexicale par contexte", Cahiers de l'analyse des données, Volume 8, Numéro 2, 1983. http://www.numdam.org/item/?id=CAD_1983__8_2_187_0
- Reinert M., "Alceste une méthodologie d'analyse des données textuelles et une application: Aurelia De Gerard De Nerval", Bulletin de Méthodologie Sociologique, Volume 26, Numéro 1, 1990. https://doi.org/10.1177/075910639002600103
- Reinert M., "Une méthode de classification des énoncés d’un corpus présentée à l’aide d’une application", Les cahiers de l’analyse des données, Tome 15, Numéro 1, 1990. http://www.numdam.org/item/?id=CAD_1990__15_1_21_0
Thanks to Pierre Ratineau, the author of Iramuteq, for providing it as free software and open source. Even if the R code has been almost entirely rewritten, it has been a precious resource to understand the algorithms.
Many thanks to Sébastien Rochette for the creation of the hex logo.
Many thanks to Florian Privé for his work on rewriting and optimizing the Rcpp code.