Rank in Network Meta-Analysis.

rankinma
Rank inN*etwork *Meta-Analysis
Chiefeng Chen, Enoch Kang, Wen-Hsuan Hou, Jin-Hua Chen, Yu-Chieh Chuang, & Edwin Chan
About rankinma
Package rankinma supports users to easily obtain and visualize various metrics of treatment ranking from network meta-analysis without major concerns regarding heterogeneity, intransitivity, and incoherence. This package not only accepts manual-prepared data set of treatment ranking metrics from users, but also can help users to gather various treatment ranking metrics in network meta-analysis.
Feature
rankinma allows users to visualize various treatment ranking metrics in network meta-analysis based either common-effect model or random-effects model no matter using frequentist or Bayesian approach. The current version includes three common metrics of treatment ranking.
- Probabilities: probabilities of every available treatment on each possible rank.
- SUCRA: the value of surface under the cumulative ranking curve using Bayesian approach.
- P-score: the value of SUCRA using frequentist approach.
Briefly, rankinma can be used for visualization of both detailed metrics of probabilities and global metrics (i.e. SUCRA and P-score). Besides, rankinma provides users multiple types of plots to illustrate aforementioned treatment ranking metrics, and current version consists of five types of plots with six sub-types.
- Beading plot: a novel graphics for displaying global metrics of treatment ranking (i.e. SUCRA and P-score) based on numeric line plot.
- Bar chart: a classic graphics for most metrics of treatment ranking (i.e. probabilities, SUCRA, and P-score), and rankinma supports two sub-type of bar chart in terms of side-by-side bar chart and cumulative bar chart.
- Line chart: a classic graphics for most metrics of treatment ranking (i.e. probabilities, SUCRA, and P-score), and rankinma supports two sub-type of line chart in terms of simple line chart (a line on a chart) and composite line chart (multiple lines on a chart).
- Heat plot: a new graphics for showing global metrics of treatment ranking (i.e. SUCRA and P-score) for each outcome, and rankinma supports to gather all heat plots of outcomes with interests on a plot.
- Spie chart: a new graphics proposed in 2020 for displaying multiple global metrics of treatment ranking (i.e. SUCRA and P-score) from outcomes with interests by each treatment, and rankinma supports to place all spie charts on a plot.
Installation
Package rankinma has been formally released on the CRAN, and users can install rankinma in R using following syntax:
install.packages("rankinma")
If users have tabulated global metrics of treatment ranking in a csv file, they can illustrate beading plot without installation of package rankinma by using online platform RankiNMA (click here).
Examples
Example 1
To depict global metrics of treatment ranking using bar chart:

Example 2
To depict probabilities of treatments on each possible rank using multi-line chart:
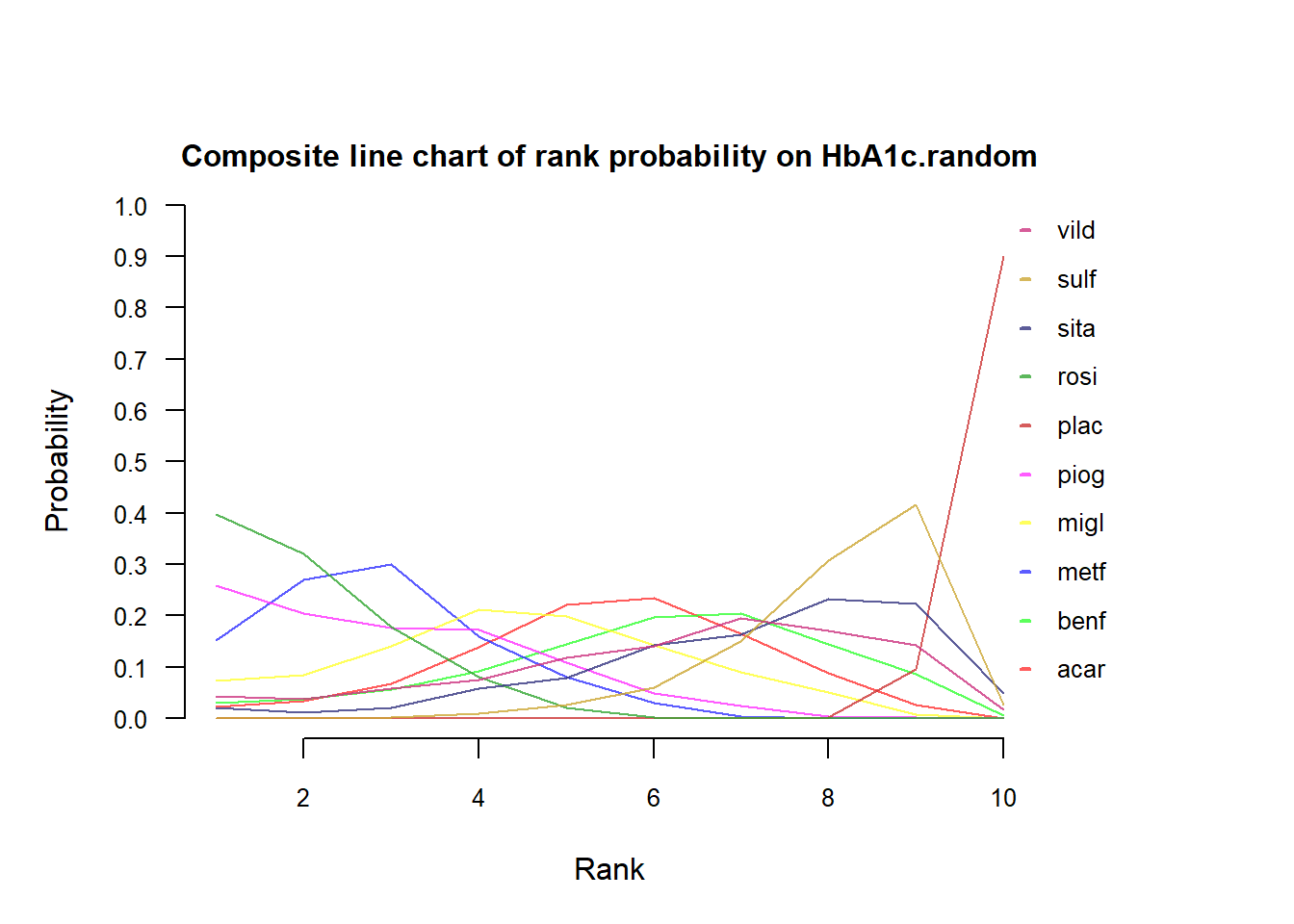
Example 3
To depict probabilities of treatments on each possible rank using stacked bar chart:

Example 4
To depict global metrics of treatment ranking using beading plot:

Coding conventions
This package is mainly written according to Google's R style. For readers, details of naming rules are listed as follows:
.R file is named using lower case with underscore "_" between words (e.g. get_metrics.R).
function is named using verb or verb with noun, and the first character of each word is written in capital letter (e.g.
GetMetrics()).object is named using noun with the first word in lower case, but the first character of rest words is written using capital letter (e.g.
dataBeads).variable is named using noun written in lower case. Words of variable name are separated by "." if a variable name consists of more than two words (e.g.
dataRankinma$metrics.name).
Version number consists of three integers with a period between them (eg. version 1.0.0).
Updating the first integer refers to a modification with new methodological impact.
Changing the second integer refers to an update with a new function without new methodological impact.
Updating the third integer refers to a modification in a function.
License
This package is licensed under the GPL-3 License.
To do list
Task force will keep update package rankinma for following issues:
- Scatter plot
- User-designed color gradient for heat plot.





