Statistical Models of Repeated Categorical Rating Data.
rater 
rater provides tools for fitting and interrogating statistical models of repeated categorical rating data. The package provides a simple interface to fit a selection of these models, with arbitrary prior parameters, using MCMC and optimisation provided by Stan. A selection of functions are also provided to plot parts of these models and extract key parameters.
Example usage:
library(rater)
fit <- rater(anesthesia, "dawid_skene") # Sampling output suppressed.
Get the posterior mean of the “pi” parameter.
point_estimate(fit, "pi")
## $pi
## [1] 0.37559632 0.40734481 0.14321934 0.07383953
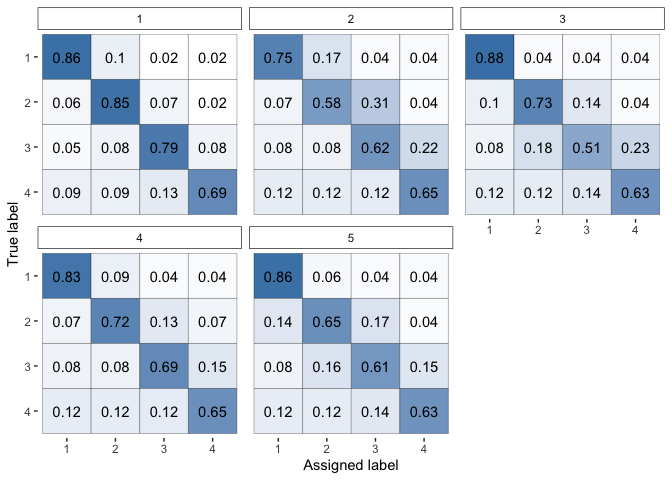
Plot the accuracy matrices of the raters.
plot(fit, "raters")
Installation
rater requires the rstan package to fit models. Detailed instructions to install rstan can be found here
CRAN
Install rater from CRAN with:
install.packages("rater")
Development
To install the development version of rater from GitHub run:
# install.packages("remotes")
remotes::install_github("jeffreypullin/rater")
Installation notes:
When installing from source, i.e. when installing the development version or installing from CRAN on Linux, the Stan models in the package will be compiled - this will lead to an install time of few minutes. Please be patient - this compilation means that no compilation is required when using the package
During compilation many warnings may be displayed in the terminal; these are harmless but impossible to suppress.