Description
Tools for Binning Data.
Description
Manually bin data using weight of evidence and information value. Includes other binning methods such as equal length, quantile and winsorized. Options for combining levels of categorical data are also available. Dummy variables can be generated based on the bins created using any of the available binning methods. References: Siddiqi, N. (2006) <doi:10.1002/9781119201731.biblio>.
README.md
rbin
Tools for binning data
Installation
# Install rbin from CRAN
install.packages("rbin")
# Or the development version from GitHub
# install.packages("devtools")
devtools::install_github("rsquaredacademy/rbin")
Addins
rbin includes two addins for manually binning data:
rbinAddin()rbinFactorAddin()
Usage
Manual Binning
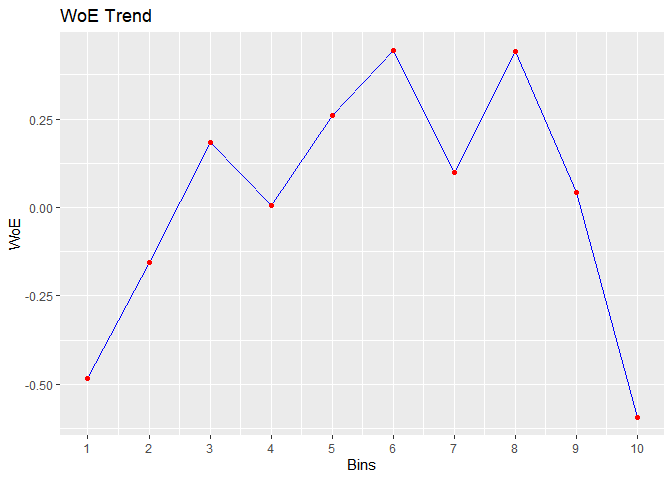
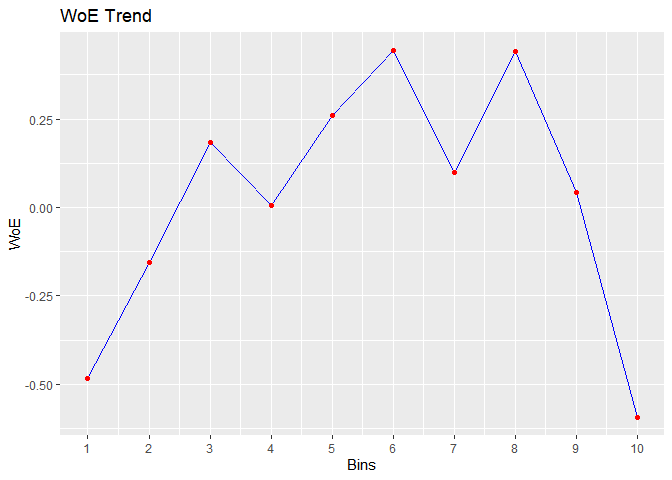
bins <- rbin_manual(mbank, y, age, c(29, 31, 34, 36, 39, 42, 46, 51, 56))
bins
#> Binning Summary
#> ---------------------------
#> Method Manual
#> Response y
#> Predictor age
#> Bins 10
#> Count 4521
#> Goods 517
#> Bads 4004
#> Entropy 0.5
#> Information Value 0.12
#>
#>
#> cut_point bin_count good bad woe iv entropy
#> 1 < 29 410 71 339 -0.483686036 2.547353e-02 0.6649069
#> 2 < 31 313 41 272 -0.154776266 1.760055e-03 0.5601482
#> 3 < 34 567 55 512 0.183985174 3.953685e-03 0.4594187
#> 4 < 36 396 45 351 0.007117468 4.425063e-06 0.5107878
#> 5 < 39 519 47 472 0.259825118 7.008270e-03 0.4383322
#> 6 < 42 431 33 398 0.442938178 1.575567e-02 0.3899626
#> 7 < 46 449 47 402 0.099298221 9.423907e-04 0.4836486
#> 8 < 51 521 40 481 0.439981550 1.881380e-02 0.3907140
#> 9 < 56 445 49 396 0.042587647 1.756117e-04 0.5002548
#> 10 >= 56 470 89 381 -0.592843261 4.564428e-02 0.7001343
# plot
plot(bins)
Combine Factor Levels
# combine levels
upper <- c("secondary", "tertiary")
out <- rbin_factor_combine(mbank, education, upper, "upper")
table(out$education)
#>
#> upper unknown primary
#> 3651 179 691
# bins
bins <- rbin_factor(out, y, education)
bins
#> Binning Summary
#> ---------------------------
#> Method Custom
#> Response y
#> Predictor education
#> Levels 3
#> Count 4521
#> Goods 517
#> Bads 4004
#> Entropy 0.51
#> Information Value 0.01
#>
#>
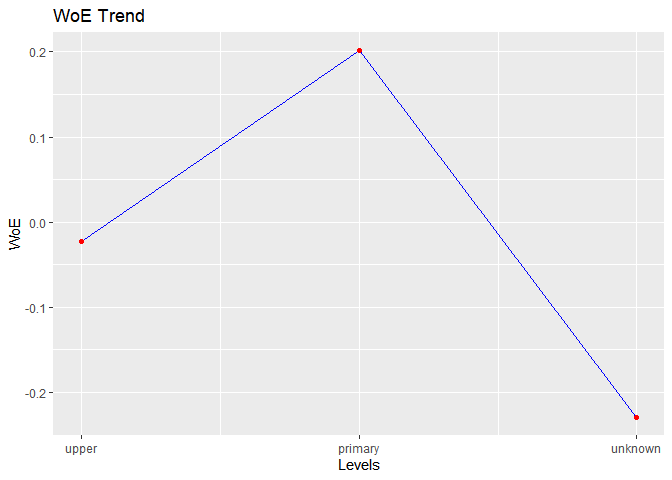
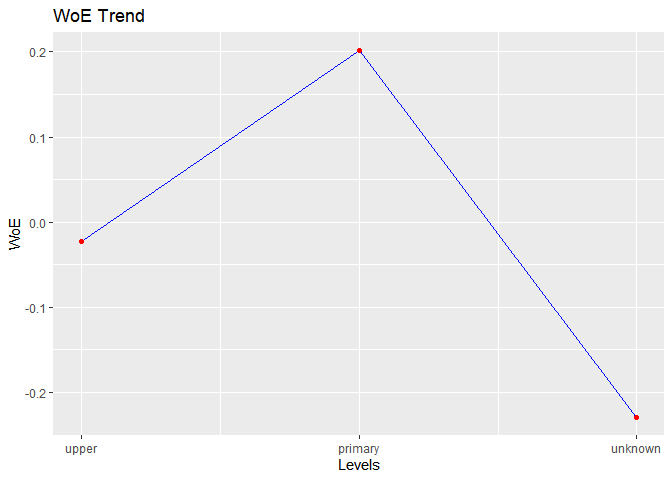
#> level bin_count good bad woe iv entropy
#> 1 upper 3651 426 3225 -0.02275738 0.0004219212 0.5197428
#> 2 primary 691 66 625 0.20109064 0.0057178780 0.4546110
#> 3 unknown 179 25 154 -0.22892949 0.0022651110 0.5833603
# plot
plot(bins)
Quantile Binning
bins <- rbin_quantiles(mbank, y, age, 10)
bins
#> Binning Summary
#> -----------------------------
#> Method Quantile
#> Response y
#> Predictor age
#> Bins 10
#> Count 4521
#> Goods 517
#> Bads 4004
#> Entropy 0.5
#> Information Value 0.12
#>
#>
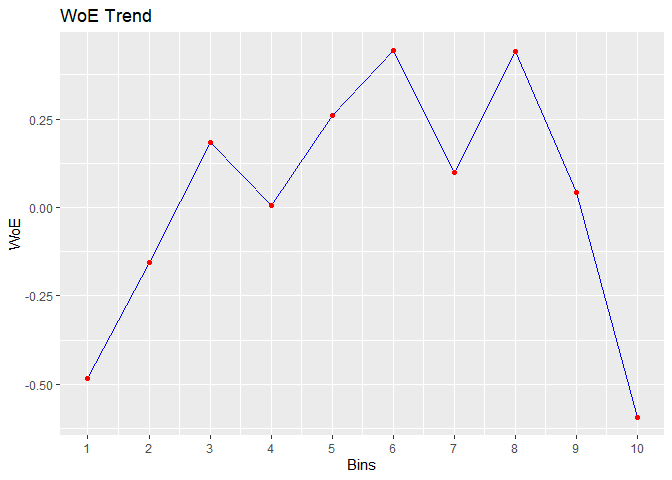
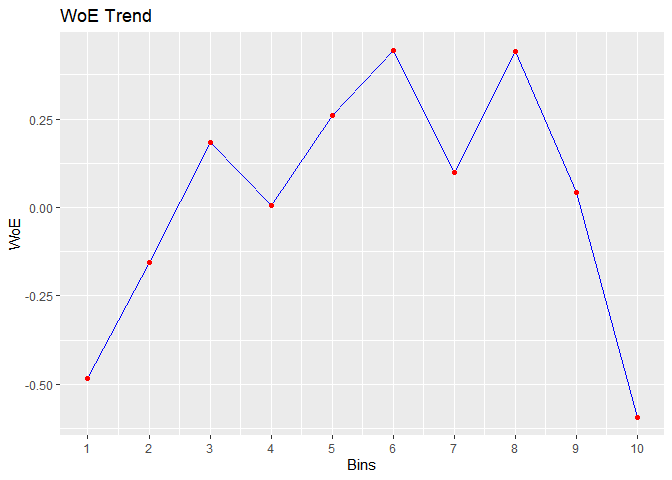
#> cut_point bin_count good bad woe iv entropy
#> 1 < 29 410 71 339 -0.483686036 2.547353e-02 0.6649069
#> 2 < 31 313 41 272 -0.154776266 1.760055e-03 0.5601482
#> 3 < 34 567 55 512 0.183985174 3.953685e-03 0.4594187
#> 4 < 36 396 45 351 0.007117468 4.425063e-06 0.5107878
#> 5 < 39 519 47 472 0.259825118 7.008270e-03 0.4383322
#> 6 < 42 431 33 398 0.442938178 1.575567e-02 0.3899626
#> 7 < 46 449 47 402 0.099298221 9.423907e-04 0.4836486
#> 8 < 51 521 40 481 0.439981550 1.881380e-02 0.3907140
#> 9 < 56 445 49 396 0.042587647 1.756117e-04 0.5002548
#> 10 >= 56 470 89 381 -0.592843261 4.564428e-02 0.7001343
# plot
plot(bins)
Winsorized Binning
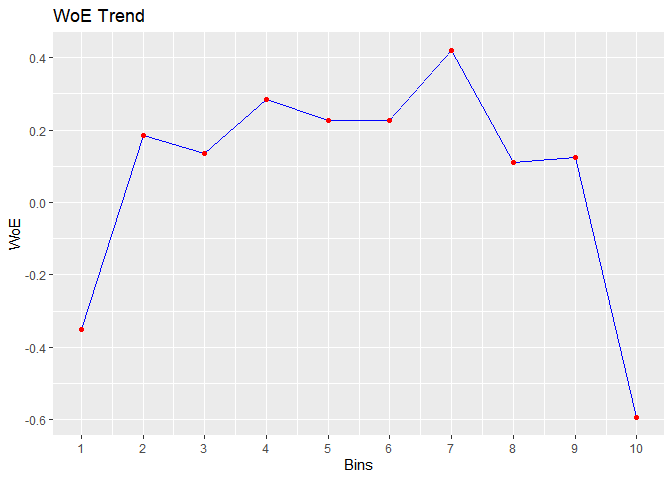
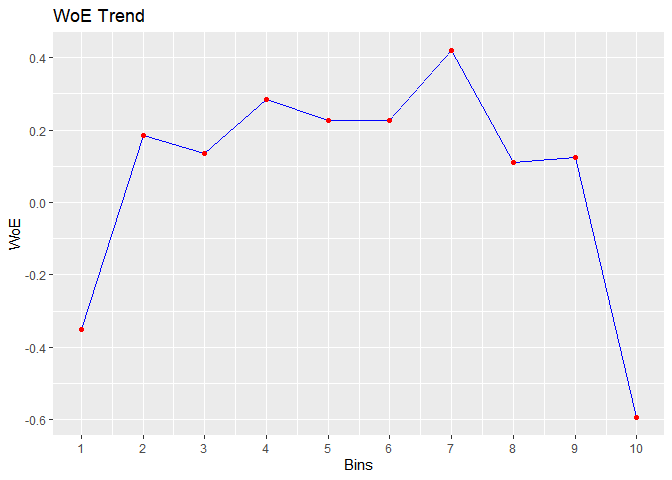
bins <- rbin_winsorize(mbank, y, age, 10, winsor_rate = 0.05)
bins
#> Binning Summary
#> ------------------------------
#> Method Winsorize
#> Response y
#> Predictor age
#> Bins 10
#> Count 4521
#> Goods 517
#> Bads 4004
#> Entropy 0.51
#> Information Value 0.1
#>
#>
#> cut_point bin_count good bad woe iv entropy
#> 1 < 30.2 723 112 611 -0.3504082 0.0224390979 0.6219926
#> 2 < 33.4 567 55 512 0.1839852 0.0039536848 0.4594187
#> 3 < 36.6 573 58 515 0.1367176 0.0022470488 0.4728562
#> 4 < 39.8 497 44 453 0.2846962 0.0079801719 0.4315480
#> 5 < 43 396 37 359 0.2253982 0.0040782670 0.4478305
#> 6 < 46.2 461 43 418 0.2272751 0.0048235624 0.4473095
#> 7 < 49.4 281 22 259 0.4187793 0.0092684760 0.3961315
#> 8 < 52.6 309 32 277 0.1112753 0.0008106706 0.4801796
#> 9 < 55.8 244 25 219 0.1231896 0.0007809490 0.4767424
#> 10 >= 55.8 470 89 381 -0.5928433 0.0456442813 0.7001343
# plot
plot(bins)
Equal Length Binning
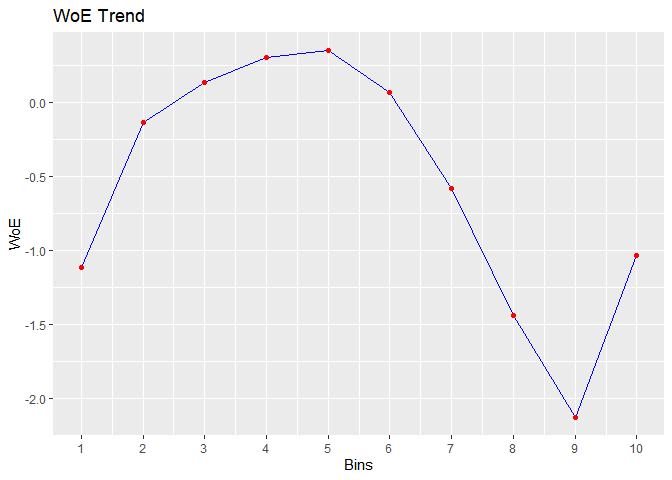
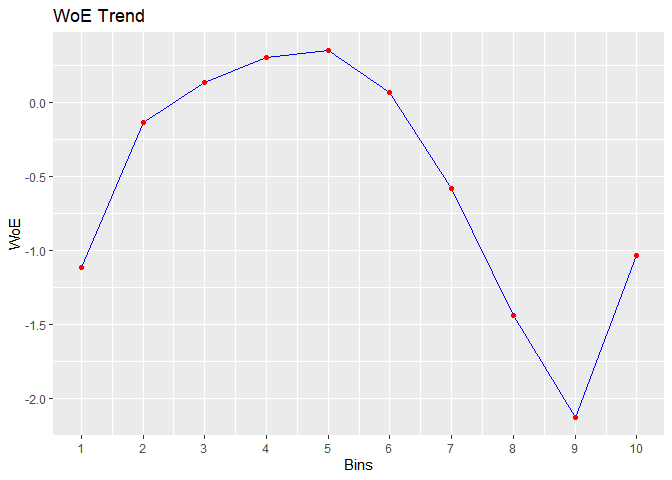
bins <- rbin_equal_length(mbank, y, age, 10)
bins
#> Binning Summary
#> ---------------------------------
#> Method Equal Length
#> Response y
#> Predictor age
#> Bins 10
#> Count 4521
#> Goods 517
#> Bads 4004
#> Entropy 0.5
#> Information Value 0.17
#>
#>
#> cut_point bin_count good bad woe iv entropy
#> 1 < 24.6 85 24 61 -1.11418623 0.0347480126 0.8586371
#> 2 < 31.2 822 106 716 -0.13676519 0.0035843196 0.5545619
#> 3 < 37.8 1133 115 1018 0.13365680 0.0042514380 0.4737339
#> 4 < 44.4 943 82 861 0.30436899 0.0171748162 0.4262287
#> 5 < 51 623 52 571 0.34913923 0.0146733167 0.4142794
#> 6 < 57.6 612 66 546 0.06595797 0.0005741022 0.4933757
#> 7 < 64.2 229 43 186 -0.58245971 0.0213871054 0.6967893
#> 8 < 70.8 34 12 22 -1.44087046 0.0255269312 0.9366674
#> 9 < 77.4 25 13 12 -2.12704897 0.0471100183 0.9988455
#> 10 >= 77.4 15 4 11 -1.03540535 0.0051663529 0.8366407
# plot
plot(bins)