Average Bioequivalence with Expanding Limits (ABEL).
replicateBE
Version 1.1.3 built 2022-05-02 with R 4.2.0 (development version not on CRAN).
Comparative BA-calculation for the EMA’s Average Bioequivalence with Expanding Limits (ABEL)
Introduction
The package provides data sets (internal .rda and in -format in /extdata/) supporting users in a black-box performance qualification (PQ) of their software installations. Users can analyze own data imported from - and Excel-files (in xlsx or the legacy xls format). The methods given by the for reference-scaling of , i.e., Average Bioequivalence with Expanding Limits (ABEL)1,2 are implemented.
Potential influence of outliers on the variability of the reference can be assessed by box plots of studentized and standardized residuals as suggested at a joint / workshop.3
Health Canada’s approach4 requiring a mixed-effects model is approximated by intra-subject contrasts.
Direct widening of the acceptance range as recommended by the Gulf Cooperation Council5 (Bahrain, Kuwait, Oman, Qatar, Saudi Arabia, United Arab Emirates) is provided as well.
In full replicate designs the variability of test and reference treatments can be assessed by swT/swR and the upper confidence limit of σwT/σwR. This was required in a pilot phase by the but lifted in 2021; reference-scaling of AUC is acceptable if the protocol is submitted to the .6
Methods
Estimation of CVwR (and CVwT in full replicate designs)
Called internally by functions method.A() and method.B(). A linear model of log-transformed pharmacokinetic (PK) responses and effects
sequence, subject(sequence), period
where all effects are fixed (i.e., by an ). Estimated by the function lm() of package stats.
modCVwR <- lm(log(PK) ~ sequence + subject%in%sequence + period,
data = data[data$treatment == "R", ])
modCVwT <- lm(log(PK) ~ sequence + subject%in%sequence + period,
data = data[data$treatment == "T", ])
Method A
Called by function method.A(). A linear model of log-transformed responses and effects
sequence, subject(sequence), period, treatment
where all effects are fixed (e.g., by an ). Estimated by the function lm() of package stats.
modA <- lm(log(PK) ~ sequence + subject%in%sequence + period + treatment,
data = data)
Method B
Called by function method.B(). A linear model of log-transformed responses and effects
sequence, subject(sequence), period, treatment
where subject(sequence) is a random effect and all others are fixed.
Three options are provided:
- Estimation by the function
lmer()of packagelmerTest.method.B(..., option = 1)employs Satterthwaite’s approximation of the degrees of freedom equivalent to SAS’DDFM=SATTERTHWAITE, Phoenix WinNonlin’sDegrees of Freedom Satterthwaite, and Stata’sdfm=Satterthwaite. Note that this is the only available approximation in SPSS.
modB <- lmer(log(PK) ~ sequence + period + treatment + (1|subject),
data = data)
- Estimation by the function
lme()of packagenlme.method.B(..., option = 2)employs degrees of freedom equivalent to SAS’DDFM=CONTAIN, Phoenix WinNonlin’sDegrees of Freedom Residual, STATISTICA’sGLM containment, and Stata’sdfm=anova. Implicitly preferred according to the ’s document and hence, the default of the function.
modB <- lme(log(PK) ~ sequence + period + treatment, random = ~1|subject,
data = data)
- Estimation by the function
lmer()of packagelmerTest.method.B(..., option = 3)employs the Kenward-Roger approximation equivalent to Stata’sdfm=Kenward Roger (EIM)and SAS’DDFM=KENWARDROGER(FIRSTORDER)i.e., based on the expected information matrix. Note that SAS withDDFM=KENWARDROGERand JMP calculate Satterthwaite’s [sic] degrees of freedom and apply the Kackar-Harville correction, i.e., based on the observed information matrix.
modB <- lmer(log(PK) ~ sequence + period + treatment + (1|subject),
data = data)
Average Bioequivalence
Called by function ABE(). The model is identical to Method A. Conventional BE limits (80.00 – 125.00%) are employed by default. Tighter limits (90.00 – 111.11%) for narrow therapeutic index drugs ( and others) or wider limits (75.00 – 133.33%) for Cmax according to the guideline of South Africa7 can be specified.
Tested designs
Four period (full) replicates
TRTR | RTRTTRRT | RTTRTTRR | RRTTTRTR | RTRT | TRRT | RTTRTRRT | RTTR | TTRR | RRTT
Three period (full) replicates
TRT | RTRTRR | RTT
Two period (full) replicate
TR | RT | TT | RR(Balaam’s design; not recommended due to poor power characteristics)
Three period (partial) replicates
TRR | RTR | RRTTRR | RTR(Extra-reference design; biased in the presence of period effects, not recommended)
Cross-validation
Details about the reference datasets:
help("data", package = "replicateBE")
?replicateBE::data
Results of the 30 reference datasets agree with ones obtained in SAS (v9.4), Phoenix WinNonlin (v6.4 – v8.3.4.295), STATISTICA (v13), SPSS (v22.0), Stata (v15.0), and JMP (v10.0.2).8
Examples
- Evaluation of the internal reference dataset 019 by Method A.
library(replicateBE) # attach the package
res <- method.A(verbose = TRUE, details = TRUE,
print = FALSE, data = rds01)
#
# Data set DS01: Method A by lm()
# ───────────────────────────────────
# Type III Analysis of Variance Table
#
# Response: log(PK)
# Df Sum Sq Mean Sq F value Pr(>F)
# sequence 1 0.0077 0.007652 0.00268 0.9588496
# period 3 0.6984 0.232784 1.45494 0.2278285
# treatment 1 1.7681 1.768098 11.05095 0.0010405
# sequence:subject 75 214.1296 2.855061 17.84467 < 2.22e-16
# Residuals 217 34.7190 0.159995
#
# treatment T – R:
# Estimate Std. Error t value Pr(>|t|)
# 0.14547400 0.04650870 3.12788000 0.00200215
# 217 Degrees of Freedom
cols <- c(12, 17:21) # extract relevant columns
# cosmetics: 2 decimal places acc. to the GL
tmp <- data.frame(as.list(sprintf("%.2f", res[cols])))
names(tmp) <- names(res)[cols]
tmp <- cbind(tmp, res[22:24]) # pass|fail
print(tmp, row.names = FALSE)
# CVwR(%) L(%) U(%) CL.lo(%) CL.hi(%) PE(%) CI GMR BE
# 46.96 71.23 140.40 107.11 124.89 115.66 pass pass pass
- The same dataset evaluated by Method B, Satterthwaite approximation of degrees of freedom.
res <- method.B(option = 1, verbose = TRUE, details = TRUE,
print = FALSE, data = rds01)
#
# Data set DS01: Method B (option = 1) by lmer()
# ──────────────────────────────────────────────
# Response: log(PK)
# Type III Analysis of Variance Table with Satterthwaite's method
# Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
# sequence 0.001917 0.001917 1 74.7208 0.01198 0.9131536
# period 0.398078 0.132693 3 217.1188 0.82881 0.4792840
# treatment 1.579332 1.579332 1 216.9386 9.86464 0.0019197
#
# treatment T – R:
# Estimate Std. Error t value Pr(>|t|)
# 0.1460900 0.0465130 3.1408000 0.0019197
# 216.939 Degrees of Freedom (equivalent to SAS’ DDFM=SATTERTHWAITE)
cols <- c(12, 17:21)
tmp <- data.frame(as.list(sprintf("%.2f", res[cols])))
names(tmp) <- names(res)[cols]
tmp <- cbind(tmp, res[22:24])
print(tmp, row.names = FALSE)
# CVwR(%) L(%) U(%) CL.lo(%) CL.hi(%) PE(%) CI GMR BE
# 46.96 71.23 140.40 107.17 124.97 115.73 pass pass pass
- The same dataset evaluated by Method B, Kenward-Roger approximation of degrees of freedom. Outlier assessment, recalculation of CVwR after exclusion of outliers, new expanded limits.
res <- method.B(option = 3, ola = TRUE, verbose = TRUE,
details = TRUE, print = FALSE, data = rds01)
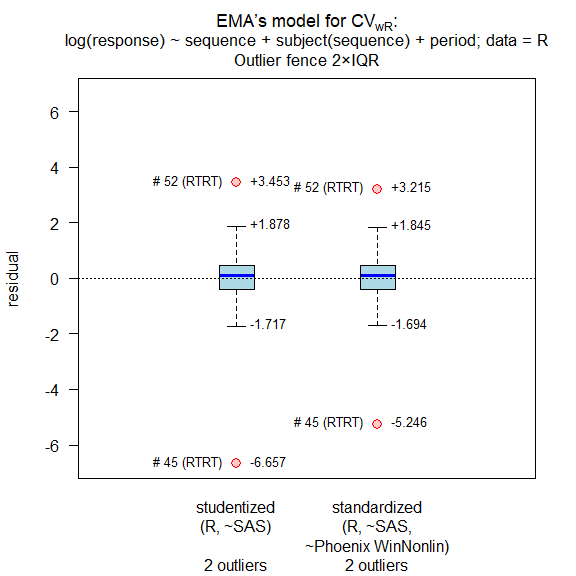
#
# Outlier analysis
# (externally) studentized residuals
# Limits (2×IQR whiskers): -1.717435, 1.877877
# Outliers:
# subject sequence stud.res
# 45 RTRT -6.656940
# 52 RTRT 3.453122
#
# standarized (internally studentized) residuals
# Limits (2×IQR whiskers): -1.69433, 1.845333
# Outliers:
# subject sequence stand.res
# 45 RTRT -5.246293
# 52 RTRT 3.214663
#
# Data set DS01: Method B (option = 3) by lmer()
# ──────────────────────────────────────────────
# Response: log(PK)
# Type III Analysis of Variance Table with Kenward-Roger's method
# Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
# sequence 0.001917 0.001917 1 74.9899 0.01198 0.9131528
# period 0.398065 0.132688 3 217.3875 0.82878 0.4792976
# treatment 1.579280 1.579280 1 217.2079 9.86432 0.0019197
#
# treatment T – R:
# Estimate Std. Error t value Pr(>|t|)
# 0.1460900 0.0465140 3.1408000 0.0019197
# 217.208 Degrees of Freedom (equivalent to Stata’s dfm=Kenward Roger EIM)
cols <- c(27, 31:32, 19:21)
tmp <- data.frame(as.list(sprintf("%.2f", res[cols])))
names(tmp) <- names(res)[cols]
tmp <- cbind(tmp, res[22:24])
print(tmp, row.names = FALSE)
# CVwR.rec(%) L.rec(%) U.rec(%) CL.lo(%) CL.hi(%) PE(%) CI GMR BE
# 32.16 78.79 126.93 107.17 124.97 115.73 pass pass pass
- The same dataset evaluated according to the conditions of the
res <- method.A(regulator = "GCC", details = TRUE,
print = FALSE, data = rds01)
cols <- c(12, 17:21)
tmp <- data.frame(as.list(sprintf("%.2f", res[cols])))
names(tmp) <- names(res)[cols]
tmp <- cbind(tmp, res[22:24])
print(tmp, row.names = FALSE)
# CVwR(%) L(%) U(%) CL.lo(%) CL.hi(%) PE(%) CI GMR BE
# 46.96 75.00 133.33 107.11 124.89 115.66 pass pass pass
- The same dataset evaluated according to the conditions of South Africa (if CVwR > 30% fixed limits 75.00 – 133.33%).
res <- ABE(theta1 = 0.75, details = TRUE,
print = FALSE, data = rds01)
tmp <- data.frame(as.list(sprintf("%.2f", res[12:17])))
names(tmp) <- names(res)[12:17]
tmp <- cbind(tmp, res[18])
print(tmp, row.names = FALSE)
# CVwR(%) BE.lo(%) BE.hi(%) CL.lo(%) CL.hi(%) PE(%) BE
# 46.96 75.00 133.33 107.11 124.89 115.66 pass
- Evaluation of the internal reference dataset 05.10 Tighter limits for the phenytoin.
res <- ABE(theta1 = 0.90, details = TRUE,
print = FALSE, data = rds05)
cols <- c(13:17)
tmp <- data.frame(as.list(sprintf("%.2f", res[cols])))
names(tmp) <- names(res)[cols]
tmp <- cbind(tmp, res[18])
print(tmp, row.names = FALSE)
# BE.lo(%) BE.hi(%) CL.lo(%) CL.hi(%) PE(%) BE
# 90.00 111.11 103.82 112.04 107.85 fail
Installation
The package requires R ≥3.5.0. However, for the Kenward-Roger approximation method.B(..., option = 3) R ≥3.6.0 is required.
- Install the released version from :
install.packages("replicateBE", repos = "https://cloud.r-project.org/")
To use the development version, please install the released version from CRAN first to get its dependencies right (readxl ≥1.0.0, PowerTOST ≥1.5.3, lmerTest, nlme, pbkrtest).
You need tools for building R packages from sources on your machine. For Windows users:
- Download Rtools from and follow the suggestions of the installer.
- Install
devtoolsand build the development version by:
install.packages("devtools", repos = "https://cloud.r-project.org/")
devtools::install_github("Helmut01/replicateBE")
Session Information
Inspect this information for reproducibility. Of particular importance are the versions of R and the packages used to create this workflow. It is considered good practice to record this information with every analysis.
options(width = 66)
print(sessionInfo(), locale = FALSE)
# R version 4.2.0 (2022-04-22 ucrt)
# Platform: x86_64-w64-mingw32/x64 (64-bit)
# Running under: Windows 10 x64 (build 22000)
#
# Matrix products: default
#
# attached base packages:
# [1] stats graphics grDevices utils datasets methods
# [7] base
#
# other attached packages:
# [1] replicateBE_1.1.3
#
# loaded via a namespace (and not attached):
# [1] tidyselect_1.1.2 xfun_0.30 purrr_0.3.4
# [4] splines_4.2.0 lmerTest_3.1-3 lattice_0.20-45
# [7] colorspace_2.0-3 vctrs_0.4.1 generics_0.1.2
# [10] htmltools_0.5.2 yaml_2.3.5 utf8_1.2.2
# [13] rlang_1.0.2 pillar_1.7.0 nloptr_2.0.0
# [16] glue_1.6.2 PowerTOST_1.5-4 readxl_1.4.0
# [19] lifecycle_1.0.1 stringr_1.4.0 munsell_0.5.0
# [22] gtable_0.3.0 cellranger_1.1.0 mvtnorm_1.1-3
# [25] evaluate_0.15 knitr_1.39 fastmap_1.1.0
# [28] parallel_4.2.0 pbkrtest_0.5.1 fansi_1.0.3
# [31] highr_0.9 broom_0.8.0 Rcpp_1.0.8.3
# [34] backports_1.4.1 scales_1.2.0 lme4_1.1-29
# [37] TeachingDemos_2.12 ggplot2_3.3.5 digest_0.6.29
# [40] stringi_1.7.6 dplyr_1.0.8 numDeriv_2016.8-1.1
# [43] grid_4.2.0 cli_3.3.0 tools_4.2.0
# [46] magrittr_2.0.3 tibble_3.1.6 tidyr_1.2.0
# [49] crayon_1.5.1 pkgconfig_2.0.3 MASS_7.3-57
# [52] ellipsis_0.3.2 Matrix_1.4-1 minqa_1.2.4
# [55] rmarkdown_2.14 rstudioapi_0.13 cubature_2.0.4.4
# [58] R6_2.5.1 boot_1.3-28 nlme_3.1-157
# [61] compiler_4.2.0
Contributors
Helmut Schütz (author) ORCID
iD
Michael Tomashevskiy (contributor)
Detlew Labes (contributor) <span style="font-size:smaller">ORCID iD
Disclaimer
Package offered for Use without any Guarantees and Absolutely No Warranty. No Liability is accepted for any Loss and Risk to Public Health Resulting from Use of this R-Code.
1.
. EMA/582648/2016.
Annex I. London. 21 September 2016.
Online↩
2.
,
.
CPMP/EWP/QWP/1401/98 Rev. 1/ Corr **. London. 20 January 2010.
Online.↩
3.
.
Revised EMA Bioequivalence Guideline. Questions & Answers. London.
June 2010.
Online↩
4. Health Canada. Guidance
Document. Conduct and Analysis of Comparative Bioavailability Studies.
Ottawa. 2018/06/08.
Online.↩
5. Executive Board of the
Health Ministers’ Council for
↩
6.
. Application of
reference-scaled criteria for AUC in bioequivalence studies conducted
for submission to
.
Geneva. 02 July 2021.
Online.↩
7.
↩
8. Schütz H, Tomashevskiy M,
Labes D, Shitova A, González-de la Parra M, Fuglsang A. Reference
Datasets for Studies in a Replicate Design Intended for Average
Bioequivalence with Expanding Limits. AAPS J. 2020; 22(2): Article 44.
doi:10.1208/s12248-020-0427-6.↩
9.
. EMA/582648/2016.
Annex II. London. 21 September 2016.
Online.↩
10. Shumaker RC, Metzler CM.
The Phenytoin Trial is a Case Study of ‘Individual’ Bioequivalence.
Drug Inf J. 1998; 32(4): 1063–72.
doi:10.1177/009286159803200426.↩









