reportRmd



The goal of reportRmd is to automate the reporting of clinical data in Rmarkdown environments. Functions include table one-style summary statistics, compilation of multiple univariate models, tidy output of multivariable models and side by side comparisons of univariate and multivariable models. Plotting functions include customisable survival curves, forest plots, and automated bivariate plots.
Installation
Installing from CRAN:
install.packages('reportRmd')
You can install the development version of reportRmd from GitHub with:
# install.packages("devtools")
devtools::install_github("biostatsPMH/reportRmd", ref="development")
New Features
- new compact summary table function
rm_compactsum - main functions are now pipeable
- new function to use variable labels in ggplots
replace_plot_labels
Documentation
Online Documentation
Examples
Summary statistics by Sex
library(reportRmd)
data("pembrolizumab")
rm_covsum(data=pembrolizumab, maincov = 'sex',
covs=c('age','pdl1','change_ctdna_group'),
show.tests=TRUE)
| Full Sample (n=94) | Female (n=58) | Male (n=36) | p-value | StatTest |
|---|
| age | | | | 0.30 | Wilcoxon Rank Sum |
| Mean (sd) | 57.9 (12.8) | 56.9 (12.6) | 59.3 (13.1) | | |
| Median (Min,Max) | 59.1 (21.1, 81.8) | 56.6 (34.1, 78.2) | 61.2 (21.1, 81.8) | | |
| pdl1 | | | | 0.76 | Wilcoxon Rank Sum |
| Mean (sd) | 13.9 (29.2) | 15.0 (30.5) | 12.1 (27.3) | | |
| Median (Min,Max) | 0 (0, 100) | 0.5 (0.0, 100.0) | 0 (0, 100) | | |
| Missing | 1 | 0 | 1 | | |
| change ctdna group | | | | 0.84 | Chi Sq |
| Decrease from baseline | 33 (45) | 19 (48) | 14 (42) | | |
| Increase from baseline | 40 (55) | 21 (52) | 19 (58) | | |
| Missing | 21 | 18 | 3 | | |
Compact Table
pembrolizumab |> rm_compactsum( grp = 'sex',
xvars=c('age','pdl1','change_ctdna_group'))
| Full Sample (n=94) | Female (n=58) | Male (n=36) | p-value | Missing |
|---|
| age | 59.1 (49.5-68.7) | 56.6 (45.8-67.8) | 61.2 (52.0-69.4) | 0.30 | 0 |
| pdl1 | 0.0 (0.0-10.0) | 0.5 (0.0-13.8) | 0.0 (0.0-4.5) | 0.76 | 1 |
| change ctdna group - Increase from
baseline | 40 (55%) | 21 (52%) | 19 (58%) | 0.84 | 21 |
Using Variable Labels
var_names <- data.frame(var=c("age","pdl1","change_ctdna_group"),
label=c('Age at study entry',
'PD L1 percent',
'ctDNA change from baseline to cycle 3'))
pembrolizumab <- set_labels(pembrolizumab,var_names)
rm_covsum(data=pembrolizumab, maincov = 'sex',
covs=c('age','pdl1','change_ctdna_group'))
| Full Sample (n=94) | Female (n=58) | Male (n=36) | p-value |
|---|
| Age at study entry | | | | 0.30 |
| Mean (sd) | 57.9 (12.8) | 56.9 (12.6) | 59.3 (13.1) | |
| Median (Min,Max) | 59.1 (21.1, 81.8) | 56.6 (34.1, 78.2) | 61.2 (21.1, 81.8) | |
| PD L1 percent | | | | 0.76 |
| Mean (sd) | 13.9 (29.2) | 15.0 (30.5) | 12.1 (27.3) | |
| Median (Min,Max) | 0 (0, 100) | 0.5 (0.0, 100.0) | 0 (0, 100) | |
| Missing | 1 | 0 | 1 | |
| ctDNA change from baseline to cycle
3 | | | | 0.84 |
| Decrease from baseline | 33 (45) | 19 (48) | 14 (42) | |
| Increase from baseline | 40 (55) | 21 (52) | 19 (58) | |
| Missing | 21 | 18 | 3 | |
Multiple Univariate Regression Analyses
rm_uvsum(data=pembrolizumab, response='orr',
covs=c('age','pdl1','change_ctdna_group'))
#> Waiting for profiling to be done...
#> Waiting for profiling to be done...
#> Waiting for profiling to be done...
| OR(95%CI) | p-value | N | Event |
|---|
| Age at study entry | 0.96 (0.91, 1.00) | 0.089 | 94 | 78 |
| PD L1 percent | 0.97 (0.95, 0.98) | \<0.001 | 93 | 77 |
| ctDNA change from baseline to cycle
3 | | 0.002 | 73 | 58 |
| Decrease from baseline | Reference | | 33 | 19 |
| Increase from baseline | 28.74 (5.20, 540.18) | | 40 | 39 |
Tidy multivariable analysis
glm_fit <- glm(orr~change_ctdna_group+pdl1+cohort,
family='binomial',
data = pembrolizumab)
rm_mvsum(glm_fit,showN=T)
| OR(95%CI) | p-value | N | Event |
|---|
| ctDNA change from baseline to cycle
3 | | 0.009 | 73 | 58 |
| Decrease from baseline | Reference | | 33 | 19 |
| Increase from baseline | 19.99 (2.08, 191.60) | | 40 | 39 |
| PD L1 percent | 0.97 (0.95, 1.00) | 0.066 | 73 | 58 |
| cohort | | | 73 | 58 |
| A | Reference | | 14 | 11 |
| B | 2.6e+07 (0e+00, Inf) | 1.00 | 11 | 11 |
| C | 4.2e+07 (0e+00, Inf) | 1.00 | 10 | 10 |
| D | 0.07 (4.2e-03, 1.09) | 0.057 | 10 | 3 |
| E | 0.44 (0.04, 5.10) | 0.51 | 28 | 23 |
Combining univariate and multivariable models
uvsumTable <- rm_uvsum(data=pembrolizumab, response='orr',
covs=c('age','sex','pdl1','change_ctdna_group'),tableOnly = TRUE)
#> Waiting for profiling to be done...
#> Waiting for profiling to be done...
#> Waiting for profiling to be done...
#> Waiting for profiling to be done...
glm_fit <- glm(orr~change_ctdna_group+pdl1,
family='binomial',
data = pembrolizumab)
mvsumTable <- rm_mvsum(glm_fit,tableOnly = TRUE)
rm_uv_mv(uvsumTable,mvsumTable)
| Unadjusted OR(95%CI) | p | Adjusted OR(95%CI) | p (adj) |
|---|
| Age at study entry | 0.96 (0.91, 1.00) | 0.089 | | |
| sex | | 0.11 | | |
| Female | Reference | | | |
| Male | 0.41 (0.13, 1.22) | | | |
| PD L1 percent | 0.97 (0.95, 0.98) | \<0.001 | 0.98 (0.96, 1.00) | 0.024 |
| ctDNA change from baseline to cycle
3 | | 0.002 | | 0.004 |
| Decrease from baseline | Reference | | Reference | |
| Increase from baseline | 28.74 (5.20, 540.18) | | 24.71 (2.87, 212.70) | |
Simple survival summary table
Shows events, median survival, survival rates at different times and the log rank test. Does not allow for covariates or strata, just simple tests between groups
rm_survsum(data=pembrolizumab,time='os_time',status='os_status',
group="cohort",survtimes=c(12,24),
# group="cohort",survtimes=seq(12,36,12),
# survtimesLbls=seq(1,3,1),
survtimesLbls=c(1,2),
survtimeunit='yr')
| Group | Events/Total | Median (95%CI) | 1yr (95% CI) | 2yr (95% CI) |
|---|
| A | 12/16 | 8.30 (4.24, Not Estimable) | 0.38 (0.20, 0.71) | 0.23 (0.09, 0.59) |
| B | 16/18 | 8.82 (4.67, 20.73) | 0.32 (0.16, 0.64) | 0.06 (9.6e-03, 0.42) |
| C | 12/18 | 17.56 (7.95, Not Estimable) | 0.61 (0.42, 0.88) | 0.44 (0.27, 0.74) |
| D | 4/12 | Not Estimable (6.44, Not Estimable) | 0.67 (0.45, 0.99) | 0.67 (0.45, 0.99) |
| E | 20/30 | 14.26 (9.69, Not Estimable) | 0.63 (0.48, 0.83) | 0.34 (0.20, 0.57) |
| | Log Rank Test | ChiSq | 11.3 on 4 df |
| | | p-value | 0.023 |
Summarise Cumulative incidence
library(survival)
data(pbc)
rm_cifsum(data=pbc,time='time',status='status',group=c('trt','sex'),
eventtimes=c(1825,3650),eventtimeunit='day')
#> 106 observations with missing data were removed.
| Strata | Event/Total | 1825day (95% CI) | 3650day (95% CI) |
|---|
| 1, f | 7/137 | 0.04 (0.01, 0.08) | 0.06 (0.03, 0.12) |
| 1, m | 3/21 | 0.10 (0.02, 0.27) | 0.16 (0.03, 0.36) |
| 2, f | 9/139 | 0.05 (0.02, 0.09) | 0.09 (0.04, 0.17) |
| 2, m | 0/15 | 0e+00 (NA, NA) | 0e+00 (NA, NA) |
| Gray’s Test | ChiSq | 3.3 on 3 df |
| | p-value | 0.35 |
Plotting survival curves
ggkmcif2(response = c('os_time','os_status'),
cov='cohort',
data=pembrolizumab)
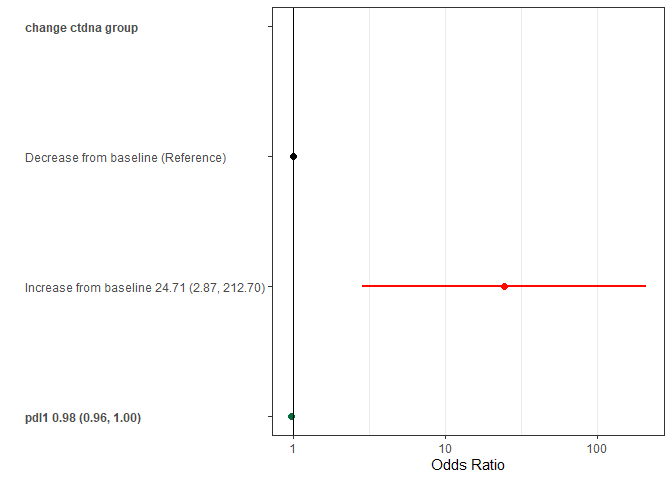
Plotting odds ratios
require(ggplot2)
#> Loading required package: ggplot2
forestplotMV(glm_fit)
#> Warning in forestplotMV(glm_fit): NAs introduced by coercion
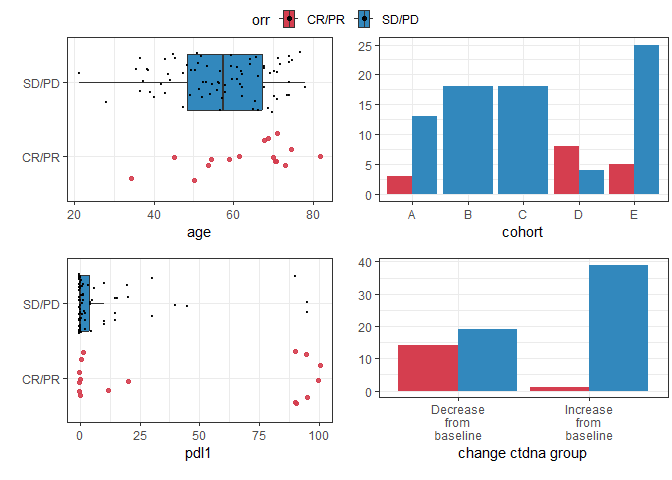
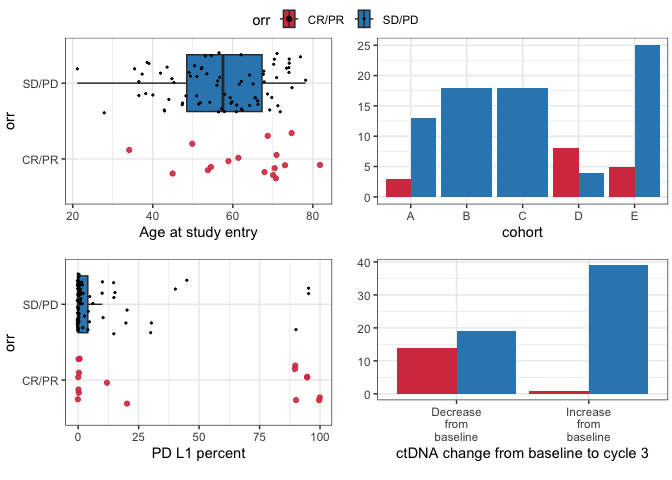
Plotting bivariate relationships
These plots are designed for quick inspection of many variables, not for publication.
require(ggplot2)
plotuv(data=pembrolizumab, response='orr',
covs=c('age','cohort','pdl1','change_ctdna_group'))
#> Boxplots not shown for categories with fewer than 20 observations.
#> Boxplots not shown for categories with fewer than 20 observations.
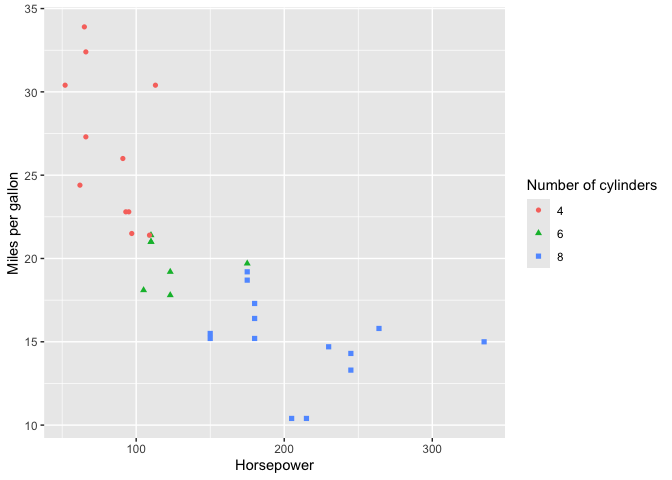
Replacing variable names with labels in ggplot
data("mtcars")
mtcars <- mtcars |>
dplyr::mutate(cyl = as.factor(cyl)) |>
set_labels(data.frame(var=c("hp","mpg","cyl"),
label=c('Horsepower',
'Miles per gallon',
'Number of cylinders')))
p <- mtcars |>
ggplot(aes(x=hp, y=mpg, color=cyl, shape=cyl)) +
geom_point()
replace_plot_labels(p)