Memory-Based Learning in Spectral Chemometrics.
resemble Memory-Based Learning in Spectral Chemometrics

Last update: 2025-10-17
Version: 2.2.5 – dstatements
Think Globally, Fit Locally (Saul and Roweis, 2003)
About
The resemble package provides high-performing functionality for data-driven modeling (including local modeling), nearest-neighbor search and orthogonal projections in spectral data.
Vignette
A new vignette for resemble explaining its core functionality is available at: https://cran.r-project.org/package=resemble/vignettes/resemble.html
Core functionality
The core functionality of the package can be summarized into the following functions:
mbl: implements memory-based learning (MBL) for modeling and predicting continuous response variables. For example, it can be used to reproduce the famous LOCAL algorithm proposed by Shenk et al. (1997). In general, this function allows you to easily customize your own MBL regression-prediction method.
dissimilarity: Computes dissimilarity matrices based on various methods (e.g. Euclidean, Mahalanobis, cosine, correlation, moving correlation, Spectral information divergence, principal components dissimilarity and partial least squares dissimilarity).
ortho_projection: A function for dimensionality reduction using either principal component analysis or partial least squares (a.k.a projection to latent structures).
search_neighbors: A function to efficiently retrieve from a reference set the k-nearest neighbors of another given dataset.
New version
During the recent lockdown we invested some of our free time to come up with a new version of our package. This new resemble 2.0 comes with MAJOR improvements and new functions! For these improvements major changes were required. The most evident changes are in the function and argument names. These have been now adapted to properly follow the tydiverse style guide. A number of changes have been implemented for the sake of computational efficiency. These changes are documented in inst\changes.md.
New interesing functions and fucntionality are also available, for example, the mbl() function now allows sample spiking, where a set of reference observations can be forced to be included in the neighborhhoods of each sample to be predicted. The serach_neighbors() function efficiently retrieves from a refence set the k-nearest neighbors of another given dataset. The dissimilarity() function computes dissimilarity matrices based on various metrics.
Installation
If you want to install the package and try its functionality, it is very simple, just type the following line in your R console:
install.packages('resemble')
If you do not have the following packages installed, it might be good to update/install them first
install.packages('Rcpp')
install.packages('RcppArmadillo')
install.packages('foreach')
install.packages('iterators')
Note: Apart from these packages we stronly recommend to download and install Rtools https://cran.r-project.org/bin/windows/Rtools/). This is important for obtaining the proper C++ toolchain that might be needed for resemble.
Then, install resemble
You can also install the development version of resemble directly from github using devtools:
devtools::install_github("l-ramirez-lopez/resemble")
NOTE: in some MAC Os it is still recommended to install gfortran and clang from here. Even for R >= 4.0. For more info, check this issue.
Example
After installing resemble you should be also able to run the following lines:
library(resemble)
library(tidyr)
library(prospectr)
data(NIRsoil)
# Proprocess the data
NIRsoil <- NIRsoil[NIRsoil$CEC %>% complete.cases(),]
wavs <- as.numeric(colnames(NIRsoil$spc))
NIRsoil$spc_p <- NIRsoil$spc %>%
standardNormalVariate() %>%
resample(wavs, seq(min(wavs), max(wavs), by = 11)) %>%
savitzkyGolay(p = 1, w = 5, m = 1)
# split into calibration/training and test
train_x <- NIRsoil$spc_p[as.logical(NIRsoil$train), ]
train_y <- NIRsoil$CEC[as.logical(NIRsoil$train)]
test_x <- NIRsoil$spc_p[!as.logical(NIRsoil$train), ]
test_y <- NIRsoil$CEC[!as.logical(NIRsoil$train)]
# Use MBL as in Ramirez-Lopez et al. (2013)
sbl <- mbl(
Xr = train_x, Yr = train_y, Xu = test_x,
k = seq(50, 130, by = 20),
method = local_fit_gpr(),
control = mbl_control(validation_type = "NNv")
)
sbl
plot(sbl)
get_predictions(sbl)
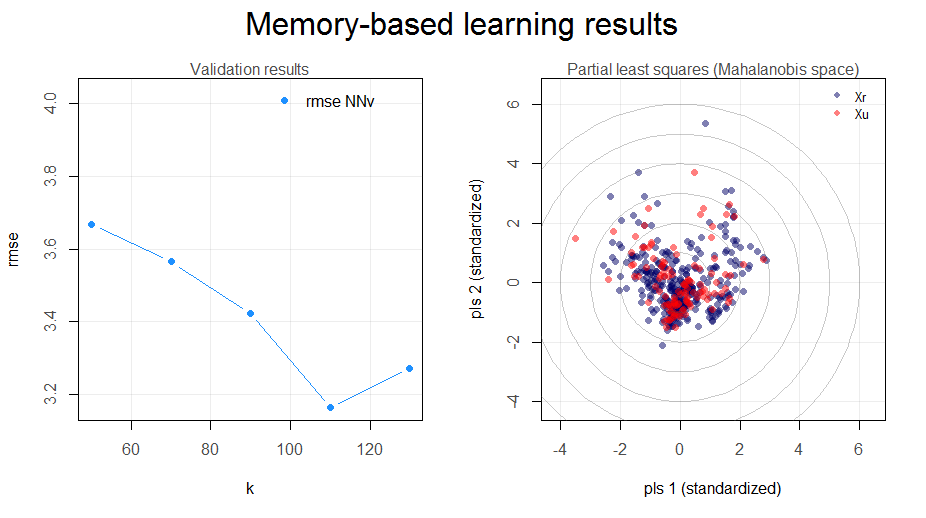
Figure 1. Standard plot of the results of the mbl function.
resemble implements functions dedicated to non-linear modelling of complex visible and infrared spectral data based on memory-based learning (MBL, a.k.a instance-based learning or local modelling in the chemometrics literature). The package also includes functions for: computing and evaluate spectral dissimilarity matrices, projecting the spectra onto low dimensional orthogonal variables, spectral neighbor search, etc.
Memory-based learning (MBL)
To expand a bit more the explanation on the mbl function, let’s define first the basic input data:
Reference (training) set: Dataset with n reference samples (e.g. spectral library) to be used in the calibration of spectral models. Xr represents the matrix of samples (containing the spectral predictor variables) and Yr represents a response variable corresponding to Xr.
Prediction set : Dataset with m samples where the response variable (Yu) is unknown. However it can be predicted by applying a spectral model (calibrated by using Xr and Yr) on the spectra of these samples (Xu).
To predict each value in Yu, the mbl function takes each sample in Xu and searches in Xr for its k-nearest neighbours (most spectrally similar samples). Then a (local) model is calibrated with these (reference) neighbours and it immediately predicts the correspondent value in Yu from Xu. In the function, the k-nearest neighbour search is performed by computing spectral dissimilarity matrices between observations. The mbl function offers the following regression options for calibrating the (local) models:
'gpr': Gaussian process with linear kernel.
'pls': Partial least squares.
'wapls': Weighted average partial least squares (Shenk et al., 1997).
Figure 2 illustrates the basic steps in MBL for a set of five observations.
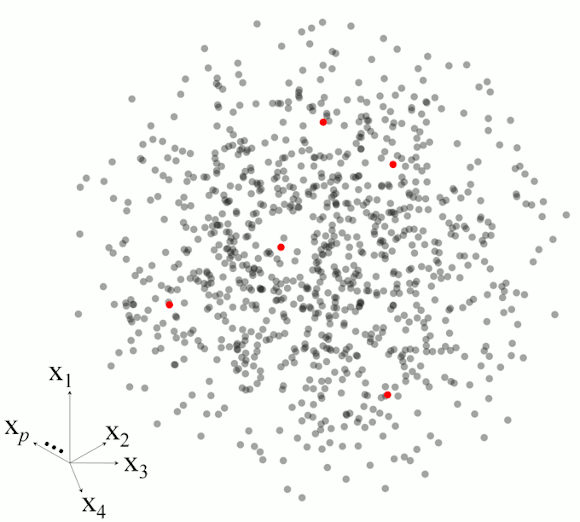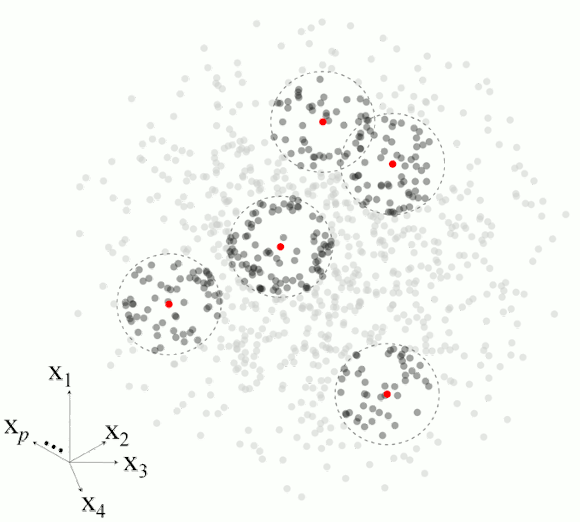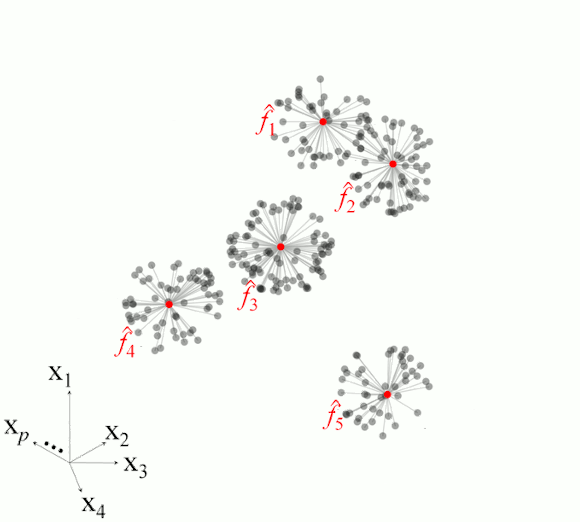
Figure 2. Example of the main steps in memory-based learning for predicting a response variable in five different observations based on set of p-dimesnional variables.
Citing the package
Simply type and you will get the info you need:
citation(package = "resemble")
News: Memory based learnig (MBL) and resemble
2025.10:Summerauer et al., 2025 used
resemblefor MBL modelling of soil physico-chemical properties from infrared spectra across tropical hillslopes in Eastern Africa. The study revealed severe soil degradation and SOC losses (up to –69 %) following deforestation, with limited recovery from Eucalyptus reforestation.2025.05:Sun and Shi, 2025 introduced a local MBL-style strategy combining spectral and geographical similarity for SOC prediction from RGB, DRS, and Sentinel-2 data; local PLSR outperformed global models and maintained accuracy across scales.
2025.03:Breure et al., 2025 published in Nature Communications:
resembleused to model NIR–soil property relations for particulate and mineral-associated organic carbon across European agricultural soils, enabling an EU-wide SOC risk index based on effective MAOC capacity and observed SOC change.2025.03:Purushothaman et al., 2025 applied MBL (
resemble) to BRDF-corrected AVIRIS-NG hyperspectral data for predicting soil properties in India; BRDF correction improved accuracy (R² up to 0.83) and reduced RMSE by up to 47 %, enabling 5 m mapping.2025.03:Kohlmann et al., 2025 fused vis–NIR and MIR with SVMR to predict total organic C, biochar C and native SOC in loess soils; cited MBL as a localisation strategy alongside spiking for site-specific transferability.
2025.02: Adam & Jackisch (LfULG report; no DOI). MIR-DRIFTS soil-C monitoring workflow, regional spectral library; compared PLSR, Cubist and MBL (
resemble). (Leave unlinked or link to an institutional page if available.)2025.01:Dai et al., 2025 used MBL and non-linear MBL (N-MBL) for POC and MAOC from VNIR in Guangdong; local
resemblemodels outperformed global ML (Cubist, PLSR, RF).2024.12:Asrat et al., 2024 MBL (
resemble) for local calibration sample selection in the Moroccan Soil Spectral Library; improved prediction of Olsen P, pH, CEC vs global PLSR.2024.11:Dai et al., 2024 used VNIR lab and in-situ spectra from wheat–rice fields in SE China (n=202) to predict SOC, POC, MAOC; PLSR/MBL performed best (lab SOC R² up to 0.91), and EPO gave modest in-situ gains, indicating feasible in-situ monitoring under low soil moisture.
2024.10:Lippolis et al., 2024 MBL (
resemble) for high-throughput NIR prediction of faba bean seed traits; compared with PLS, Elastic Net and Bayes-B.2024.09:Barbetti et al., 2024 MBL to detect SOC changes in long-term experiments using vis–NIR; R² up to 0.91; compared with Cubist, SVM and RF.
2024.09:Moloney et al., 2024 MBL with the NZ Soil Spectral Library for SOC, TN and pH in Tonga; supplementing with small local sets markedly improved accuracy.
2024.08:Sherman et al., 2024 MBL with NIR for non-destructive characterisation of silicate materials (geoarchaeology).
2023.11:Wang et al., 2023 N-MBL (MBL + RF within local fitting) improved regional vis–NIR models for SOM, TN, TP vs MBL/PLSR/Cubist/SVM/CNN.
2023.04:Zhao et al., 2023 used MBL (
resemble) + compositional data analysis to quantify soil properties relevant to SOC biogeochemical cycles from IR spectra.2022:Sanderman et al., 2022 evaluated transferability of large MIR spectral databases across instruments; MBL via
resemble.2022:Dangal et al., 2022 improved soil carbon estimates;
resembleused within the workflow.2022.01:Ng et al., 2022 showed that “spiking” regional Vis–NIR libraries with local samples does not outperform localized models. Using Australian soils, they found that memory-based learning (MBL) yield better local SOC predictions. They used
resemble.2021.12:Yu et al., 2022 MBL with External Parameter Orthogonalization for field prediction of soil properties.
2021.10:Ramirez-Lopez et al., 2021 MBL to predict soil properties in Africa.
2020.08: Charlotte Rivard’s MIR MBL tutorial: https://whrc.github.io/Soil-Predictions-MIR/
2020.04:Tsakiridis et al., 2020 used optimal principal-components dissimilarity with CNNs for simultaneous vis–NIR prediction.
2019.04:Tziolas et al., 2019 improved MBL for quantitative soil predictions using NIR + geographic information.
2019.03 & 2019.08:Tsakiridis et al., 2019a, 2019b compared ML methods for predictive soil spectroscopy; MBL (
resemble) highly competitive.2020.01:Sanderman et al., 2020 MIR spectroscopy for prediction of soil health indicators in the United States; MBL and Cubist excelled.
2019.03:Ramirez-Lopez et al., 2019 used MBL in digital soil mapping (farm-scale vis–NIR); here MBL removed local calibration outliers.
2019.01:Dangal et al., 2019
resemblefor MIR modelling from a continental US soil spectral library.2019.03:Jaconi et al., 2019 MBL (
resemble) for national-scale NIR texture predictions in Germany.2018.12:Hong/Chen et al., 2019 fractional-order derivatives + MBL (
resemble) improved NIR prediction of SOM in China.2018.11: Rivera et al., 2018 — Frontiers in Plant Science (common beans). (If you keep this, link the article’s DOI from Frontiers; avoid Google Scholar.)
2018.07:Gholizadeh et al., 2018 soil-carbon modelling with
resemble.2018.01:Dotto et al., 2018 MBL (
resemble) for SOC prediction in Brazil.2017.11: Kopf et al., 2017 (KIT Scientific Publishing, OCM 2017 proceedings). (Conference chapter/book—use the publisher page if you want a link; the Google Books preview can trigger CRAN URL notes.)
2016.05:Clairotte et al., 2016 national-scale SOC from diffuse IR spectroscopy.
2016.04:Viscarra Rossel et al., 2016 memory-based learning applied to predict soil properties.
2016.04: Blog examples on
resemble: http://nir-quimiometria.blogspot.com/ (keep as plain HTTP; it’s fine for CRAN).2016.02:
resembleon CRAN: https://CRAN.R-project.org/package=resemble2016.01:
resemble1.2 submitted to CRAN; development on GitHub.2015.11: 1.2.0 prerelease; core routines moved to C++ via Rcpp for speed.
2015.11: 1.1.3 never released to CRAN due to performance refactor.
2014.10: 1.1.3 prerelease (website); pending CRAN at the time.
2014.06: Video on local calibrations: https://www.youtube.com/watch?v=7sCIEeNehgE
2014.03: First CRAN release of
resemble.
Other R’elated stuff
- Check our other project called
prospectr. - Check this presentation in which we used the resemble package to predict soil attributes from large scale soil spectral libraries.https://www.fao.org/fileadmin/user_upload/GSP/docs/Spectroscopy_dec13/SSW2013_f.pdf
Bug report and development version
You can send an e-mail to the package maintainer ([email protected]) or create an issue on github.
References
Lobsey, C. R., Viscarra Rossel, R. A., Roudier, P., & Hedley, C. B. 2017. rs-local data-mines information from spectral libraries to improve local calibrations. European Journal of Soil Science, 68(6), 840-852.
Ramirez-Lopez, L., Behrens, T., Schmidt, K., Stevens, A., Dematte, J.A.M., Scholten, T. 2013. The spectrum-based learner: A new local approach for modeling soil vis-NIR spectra of complex data sets. Geoderma 195-196, 268-279.
Saul, L. K., & Roweis, S. T. 2003. Think globally, fit locally: unsupervised learning of low dimensional manifolds. Journal of machine learning research, 4(Jun), 119-155.
Shenk, J., Westerhaus, M., and Berzaghi, P. 1997. Investigation of a LOCAL calibration procedure for near infrared instruments. Journal of Near Infrared Spectroscopy, 5, 223-232.