Create and Query a Local Copy of 'GenBank' in R.
Locally query GenBank 
NOTE: Starting with v2.0.0, the database backend changed from MonetDBLite to duckdb. Because of this change, restez v2.0.0 or higher is not compatible with databases built with previous versions of restez.
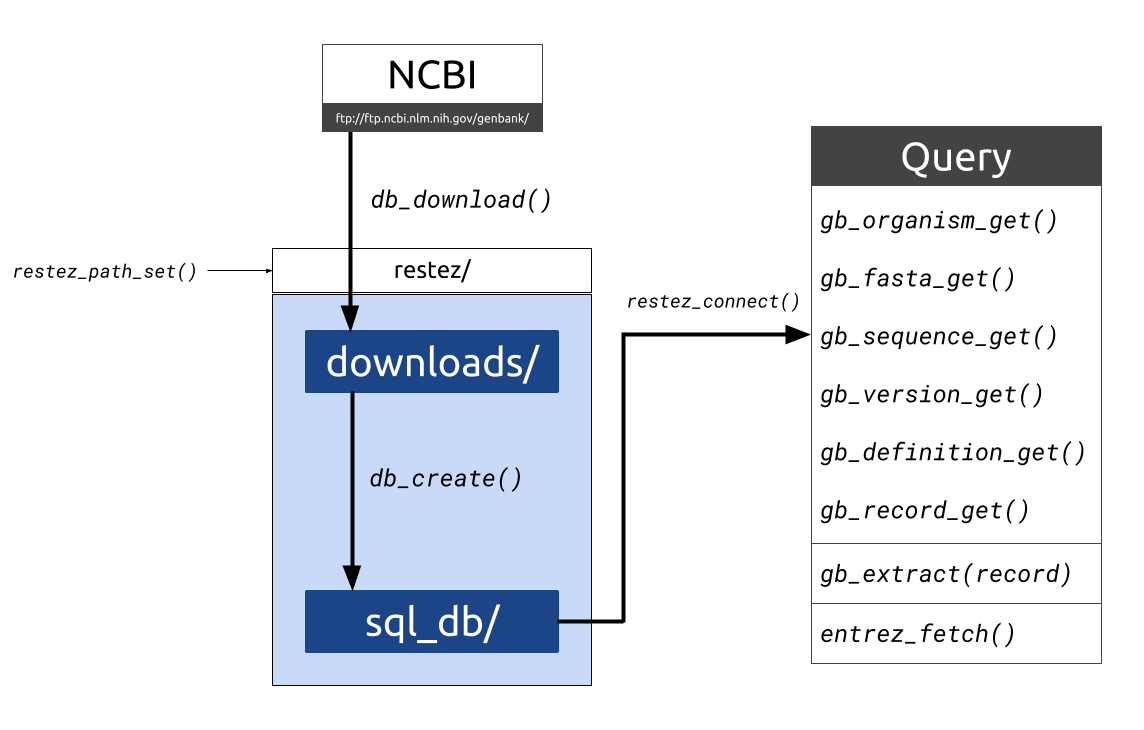
Download parts of NCBI’s GenBank to a local folder and create a simple SQL-like database. Use ‘get’ tools to query the database by accession IDs. rentrez wrappers are available, so that if sequences are not available locally they can be searched for online through Entrez.
See the detailed tutorials for more information.
Introduction
Vous entrez, vous rentrez et, maintenant, vous …. restez!
Downloading sequences and sequence information from GenBank and related NCBI taxonomic databases is often performed via the NCBI API, Entrez. Entrez, however, has a limit on the number of requests and downloading large amounts of sequence data in this way can be inefficient. For programmatic situations where multiple Entrez calls are made, downloading may take days, weeks or even months.
This package aims to make sequence retrieval more efficient by allowing a user to download large sections of the GenBank database to their local machine and query this local database either through package specific functions or Entrez wrappers. This process is more efficient as GenBank downloads are made via NCBI’s FTP using compressed sequence files. With a good internet connection and a middle-of-the-road computer, a database comprising 20 GB of sequence information can be generated in less than 10 minutes.
Installation
Install from CRAN:
install.packages("restez")
Or install the development version from r-universe:
install.packages("restez", repos = "https://ropensci.r-universe.dev")
Or install the development version from GitHub (requires installing the remotes package first):
# install.packages("remotes")
remotes::install_github("ropensci/restez")
Quick Examples
For more detailed information on the package’s functions and detailed guides on downloading, constructing and querying a database, see the detailed tutorials.
Setup
# Warning: running these examples may take a few minutes
library(restez)
# choose a location to store GenBank files
restez_path_set(rstz_pth)
# Run the download function
db_download()
# after download, create the local database
db_create()
Query
# for reproducibility
set.seed(12345)
# get a random accession ID from the database
id <- sample(list_db_ids(), 1)
#> Warning in list_db_ids(): Number of ids returned was limited to [100].
#> Set `n=NULL` to return all ids.
# you can extract:
# sequences
seq <- gb_sequence_get(id)[[1]]
str(seq)
#> chr "ACCGTTTTGACAGGTAACGTGAAAGCTCTTGGCAACGGGTCTTGATACCGAGTCGGGATCGGTAGTTGTTGCTTTGTTCGTTCACGATTTAAGGTCAACCTTAGCCTTGAGTTTTTCCAAGTAGT"
# definitions
def <- gb_definition_get(id)[[1]]
print(def)
#> [1] "Unidentified RNA clone M33.7"
# organisms
org <- gb_organism_get(id)[[1]]
print(org)
#> [1] "unidentified"
# or whole records
rec <- gb_record_get(id)[[1]]
cat(rec)
#> LOCUS AF040767 125 bp RNA linear UNA 06-MAR-1998
#> DEFINITION Unidentified RNA clone M33.7.
#> ACCESSION AF040767
#> VERSION AF040767.1
#> KEYWORDS .
#> SOURCE unidentified
#> ORGANISM unidentified
#> unclassified sequences.
#> REFERENCE 1 (bases 1 to 125)
#> AUTHORS Pan,W.S., Ji,X.Y., Wang,H.T., Tian,K.G. and Yu,X.L.
#> TITLE RNA from plasma of Rhesus monkey(NO.33) which was infected by a
#> certain patient's serum
#> JOURNAL Unpublished
#> REFERENCE 2 (bases 1 to 125)
#> AUTHORS Pan,W.S., Ji,X.Y., Wang,H.T., Tian,K.G. and Yu,X.L.
#> TITLE Direct Submission
#> JOURNAL Submitted (31-DEC-1997) Department of Applied Molecular Biology,
#> Microbiology & Epidemiology Institution, 20 Dongdajie Street,
#> Fengtai, Beijing 100071, China
#> FEATURES Location/Qualifiers
#> source 1..125
#> /organism="unidentified"
#> /mol_type="genomic RNA"
#> /db_xref="taxon:32644"
#> /clone="M33.7"
#> /note="from the plasma of Rhesus monkey which was infected
#> by plasma of a human patient"
#> ORIGIN
#> 1 accgttttga caggtaacgt gaaagctctt ggcaacgggt cttgataccg agtcgggatc
#> 61 ggtagttgtt gctttgttcg ttcacgattt aaggtcaacc ttagccttga gtttttccaa
#> 121 gtagt
#> //
Entrez wrappers
# use the entrez_* wrappers to access GB data
res <- entrez_fetch(db = 'nucleotide', id = id, rettype = 'fasta')
cat(res)
#> >AF040767.1 Unidentified RNA clone M33.7
#> ACCGTTTTGACAGGTAACGTGAAAGCTCTTGGCAACGGGTCTTGATACCGAGTCGGGATCGGTAGTTGTT
#> GCTTTGTTCGTTCACGATTTAAGGTCAACCTTAGCCTTGAGTTTTTCCAAGTAGT
# if the id is not in the local database
# these wrappers will search online via the rentrez package
res <- entrez_fetch(db = 'nucleotide', id = c('S71333.1', id),
rettype = 'fasta')
#> [1] id(s) are unavailable locally, searching online.
cat(res)
#> >AF040767.1 Unidentified RNA clone M33.7
#> ACCGTTTTGACAGGTAACGTGAAAGCTCTTGGCAACGGGTCTTGATACCGAGTCGGGATCGGTAGTTGTT
#> GCTTTGTTCGTTCACGATTTAAGGTCAACCTTAGCCTTGAGTTTTTCCAAGTAGT
#>
#> >S71333.1 alpha 1,3 galactosyltransferase [New World monkeys, mermoset lymphoid cell line B95.8, mRNA Partial, 1131 nt]
#> ATGAATGTCAAAGGAAAAGTAATTCTGTCGATGCTGGTTGTCTCAACTGTGATTGTTGTGTTTTGGGAAT
#> ATATCAACAGCCCAGAAGGCTCTTTCTTGTGGATATATCACTCAAAGAACCCAGAAGTTGATGACAGCAG
#> TGCTCAGAAGGACTGGTGGTTTCCTGGCTGGTTTAACAATGGGATCCACAATTATCAACAAGAGGAAGAA
#> GACACAGACAAAGAAAAAGGAAGAGAGGAGGAACAAAAAAAGGAAGATGACACAACAGAGCTTCGGCTAT
#> GGGACTGGTTTAATCCAAAGAAACGCCCAGAGGTTATGACAGTGACCCAATGGAAGGCGCCGGTTGTGTG
#> GGAAGGCACTTACAACAAAGCCATCCTAGAAAATTATTATGCCAAACAGAAAATTACCGTGGGGTTGACG
#> GTTTTTGCTATTGGAAGATATATTGAGCATTACTTGGAGGAGTTCGTAACATCTGCTAATAGGTACTTCA
#> TGGTCGGCCACAAAGTCATATTTTATGTCATGGTGGATGATGTCTCCAAGGCGCCGTTTATAGAGCTGGG
#> TCCTCTGCGTTCCTTCAAAGTGTTTGAGGTCAAGCCAGAGAAGAGGTGGCAAGACATCAGCATGATGCGT
#> ATGAAGACCATCGGGGAGCACATCTTGGCCCACATCCAACACGAGGTTGACTTCCTCTTCTGCATGGATG
#> TGGACCAGGTCTTCCAAGACCATTTTGGGGTAGAGACCCTGGGCCAGTCGGTGGCTCAGCTACAGGCCTG
#> GTGGTACAAGGCAGATCCTGATGACTTTACCTATGAGAGGCGGAAAGAGTCGGCAGCATATATTCCATTT
#> GGCCAGGGGGATTTTTATTACCATGCAGCCATTTTTGGAGGAACACCGATTCAGGTTCTCAACATCACCC
#> AGGAGTGCTTTAAGGGAATCCTCCTGGACAAGAAAAATGACATAGAAGCCGAGTGGCATGATGAAAGCCA
#> CCTAAACAAGTATTTCCTTCTCAACAAACCCTCTAAAATCTTATCTCCAGAATACTGCTGGGATTATCAT
#> ATAGGCCTGCCTTCAGATATTAAAACTGTCAAGCTATCATGGCAAACAAAAGAGTATAATTTGGTTAGAA
#> AGAATGTCTGA
Contributing
Want to contribute? Check the contributing page.
Licence
MIT
Citation
Bennett et al. (2018). restez: Create and Query a Local Copy of GenBank in R. Journal of Open Source Software, 3(31), 1102. https://doi.org/10.21105/joss.01102
References
Benson, D. A., Karsch-Mizrachi, I., Clark, K., Lipman, D. J., Ostell, J., & Sayers, E. W. (2012). GenBank. Nucleic Acids Research, 40(Database issue), D48–D53. https://doi.org/10.1093/nar/gkr1202
Winter DJ. (2017) rentrez: An R package for the NCBI eUtils API. PeerJ Preprints 5:e3179v2 https://doi.org/10.7287/peerj.preprints.3179v2
Maintainer
This package previously developed and maintained by Dom Bennett.
