Neutral Landscape Generator with Targets on Landscape Indices.
rflsgen 
Neutral Landscape Generator with Targets on Landscape Indices
Logo by Camille Salmon
rflsgen is the R distribution of flsgen, a neutral landscape generator that allows users to set targets on landscape indices. It first relies on Choco-solver to identify landscape structure satisfying user targets, then uses a stochastic algorithm to produce landscape rasters.
For more details and tutorials, please visit rflsgen's website: https://dimitri-justeau.github.io/rflsgen/
Download and installation
Java 8+ must be installed in your system to run rflsgen. Download and installation instructions for Java are available here: https://www.oracle.com/java/technologies/javase-downloads.html, or here: https://openjdk.java.net/install/. To provide an R interface to flsgen, which is written in Java, rflsgen relies on rJava, which is a dependency of rflsgen and will be installed automatically. If you have any trouble during the installation of rflsgen due to rJava, please refer to rJava's documentation: https://rforge.net/rJava/index.html.
rflsgen is available on CRAN, so you can install it from R using the following command:
install.packages("rflsgen")
library(rflsgen)
To install rflsgen from Github, you can use the devtools library (https://www.r-project.org/nosvn/pandoc/devtools.html) and use the following commands in R:
devtools::install_github("dimitri-justeau/rflsgen")
library(rflsgen)
Quickstart
Generating a fractal terrain raster
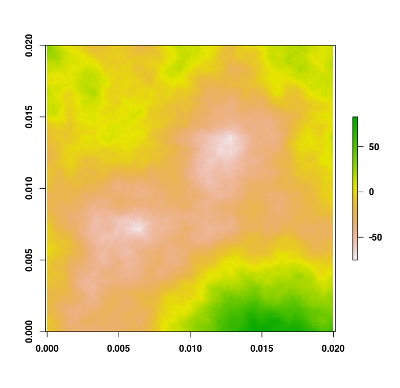
You can easily generate a fractal terrain raster using the flsgen_terrain function. For example, if you want to generate a 200x200 fractal terrain with default parameters, use the following command:
terrain <- flsgen_terrain(200, 200)
plot(terrain)
Generating landscape structures from targets
Say that we want to generate a landscape structure for a 200x200 landscape containing three landscape classes (plus a non focal class), with the following user targets:
| Class | NP | AREA | CA | MESH | PLAND |
|---|---|---|---|---|---|
| 0 | [1, 10] | [300, 4000] | [1000, 5000] | 225 | - |
| 1 | [2, 8] | [200, 4000] | - | - | 40% |
| 2 | [5, 7] | [800, 1200] | - | - | - |
The first possibility is to create a JSON file (e.g. target.json) describing these targets (note that you can also store this json in a string variable):
{
"nbRows" : 200,
"nbCols" : 200,
"classes" : [
{
"name" : "Class A",
"NP" : [1, 10],
"AREA" : [300, 4000],
"CA" : [1000, 5000],
"MESH" : [225, 225]
},
{
"name" : "Class B",
"NP" : [2, 8],
"AREA" : [200, 4000],
"PLAND" : [40, 40]
},
{
"name" : "Class C",
"NP" : [5, 7],
"AREA" : [800, 1200]
}
]
}
The second possibility is to use rflsgen helper functions:
cls_a <- flsgen_create_class_targets(
"Class A",
NP = c(1, 10),
AREA = c(300, 4000),
CA = c(1000, 5000),
MESH = c(225, 225)
)
cls_b <- flsgen_create_class_targets(
"Class B",
NP = c(2, 8),
AREA = c(200, 4000),
PLAND = c(40, 40)
)
cls_c <- flsgen_create_class_targets(
"Class C",
NP = c(5, 7),
AREA = c(800, 1200)
)
ls_targets <- flsgen_create_landscape_targets(
nb_rows = 200,
nb_cols = 200,
classes = list(cls_a, cls_b, cls_c)
)
Using the flsgen_structure function, you can generate a non-spatially-explicit landscape structure:
structure <- flsgen_structure(targets_file = "examples/targets.json")
or
structure <- flsgen_structure(ls_targets)
The result is a JSON-formatted string that contains the generated structure. It can be easily converted into a data frame with a dedicated library such as jsonlite.
Generating landscape rasters from landscape structures
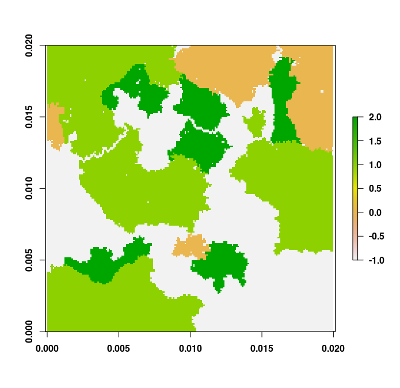
Now, let's generate a landscape raster from the previously generated structure. To do so, we use the flsgen_generate function to generate a landscape raster from the previously generated landscape structure:
landscape <- flsgen_generate(structure_str = structure)
plot(landscape)
Masking
It is possible to use a mask raster, whose NO_DATA cell will be unavailable for both focal and non-focal classes. To do so, instead of specifying the number of rows and columns in the targets, specify the mask raster with the maskRasterPath key:
{
"maskRasterPath": "mask.tif"
"classes" : [
{
"name" : "Class A",
"NP" : [2, 30],
"AREA" : [200, 4000],
"PLAND" : [40, 40]
}
]
}
or:
cls_a <- flsgen_create_class_targets(
"Class A",
NP = c(2, 30),
AREA = c(200, 4000),
PLAND = c(40, 40)
)
ls_targets <- flsgen_create_landscape_targets(
mask_raster = "mask.tif",
classes = list(cls_a)
)
Extracting structures from existing landscapes
Instead of generating landscape structure from targets, it is also possible to extract existing structures from real landscapes and use them to recreate real composition patterns. To do so, simply use the flsgen_extract_structure_from_raster function, indicating the raster values of focal classes:
struct <- flsgen_extract_structure_from_raster("existing_raster.tif", focal_classes=c(0, 1))
Citation
Please cite the rflsgen R package when using it in publications.
Justeau-Allaire, D., Blanchard, G., Ibanez, T., Lorca, X., Vieilledent, G. & Birnbaum, P. (2022). Fragmented landscape generator (flsgen): A neutral landscape generator with control of landscape structure and fragmentation indices. Methods in Ecology and Evolution, 00, 1– 9. https://doi.org/10.1111/2041-210X.13859
