G-Computation to Estimate Interpretable Epidemiological Effects.
riskCommunicator
An R package for estimating risk differences and relative risk measures.
The riskCommunicator package facilitates the estimation of common epidemiological effect measures that are relevant to public health, but that are often not trivial to obtain from common regression models, like logistic regression. In particular, riskCommunicator estimates risk and rate differences, in addition to risk and rate ratios. The package estimates these effects using g-computation with the appropriate parametric model depending on the outcome (logistic regression for binary outcomes, Poisson regression for rate or count outcomes, negative binomial regression for overdispersed rate or count outcomes, and linear regression for continuous outcomes). Therefore, the package can handle binary, rate, count, and continuous outcomes and allows for dichotomous, categorical (>2 categories), or continuous exposure variables. Additional features include estimation of effects stratified by subgroup and adjustment of standard errors for clustering. Confidence intervals are constructed by bootstrap at the individual or cluster level, as appropriate.
This package operationalizes g-computation, which has not been widely adopted due to computational complexity, in an easy-to-use implementation tool to increase the reporting of more interpretable epidemiological results. To make the package accessible to a broad range of health researchers, our goal was to design a function that was as straightforward as the standard logistic regression functions in R (e.g. glm) and that would require little to no expertise in causal inference methods or advanced coding.
Installation
You can install the released version of riskCommunicator from CRAN with:
install.packages("riskCommunicator")
The development version is available as a source package through GitHub. Installation requires the ability to compile R packages. This means that R and the R tool-chain must be installed, which requires the Xcode command-line tools on Mac and Rtools on Windows.
The easiest source installation method uses the devtools package:
# install.packages("devtools")
devtools::install_github("jgrembi/riskCommunicator")
Other Resources
Bugs and difficulties in using riskCommunicator are welcome on the issue tracker.
Planned feature improvements are also publicly catalogued on the “Issues” page for riskCommunicator: https://github.com/jgrembi/riskCommunicator/issues
Example
This is a basic example which shows you how to answer the following question: What is the effect of obesity on the 24-year risk of cardiovascular disease or death due to any cause?
In this example, we specify obesity as a categorical variable (bmicat coding: 0 = normal weight; 1=underweight; 2=overweight; 3=obese)
library(riskCommunicator)
library(ggplot2)
library(tidyverse)
#> ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.1 ──
#> ✔ tibble 3.1.7 ✔ dplyr 1.0.9
#> ✔ tidyr 1.2.0 ✔ stringr 1.4.0
#> ✔ readr 2.1.2 ✔ forcats 0.5.1
#> ✔ purrr 0.3.4
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
## basic example code
data(cvdd)
set.seed(345)
bmi.results <- gComp(data = cvdd, Y = "cvd_dth", X = "bmicat", Z = c("AGE", "SEX", "DIABETES", "CURSMOKE", "PREVHYP"), outcome.type = "binary", R = 200)
summary(bmi.results)
#> Formula:
#> cvd_dth ~ bmicat + AGE + SEX + DIABETES + CURSMOKE + PREVHYP
#>
#> Family: binomial
#> Link function: logit
#>
#> Contrast: bmicat1 v. bmicat0 bmicat2 v. bmicat0 bmicat3 v. bmicat0
#>
#> Parameter estimates:
#> bmicat1_v._bmicat0 Estimate (95% CI)
#> Risk Difference 0.042 (-0.082, 0.178)
#> Risk Ratio 1.106 (0.792, 1.454)
#> Odds Ratio 1.248 (0.631, 2.506)
#> Number needed to treat/harm 23.846
#> bmicat2_v._bmicat0 Estimate (95% CI)
#> Risk Difference 0.017 (-0.005, 0.049)
#> Risk Ratio 1.044 (0.988, 1.126)
#> Odds Ratio 1.097 (0.973, 1.297)
#> Number needed to treat/harm 57.405
#> bmicat3_v._bmicat0 Estimate (95% CI)
#> Risk Difference 0.093 (0.056, 0.136)
#> Risk Ratio 1.235 (1.143, 1.354)
#> Odds Ratio 1.628 (1.348, 2.037)
#> Number needed to treat/harm 10.733
#>
#> Underlying glm:
#> Call: stats::glm(formula = formula, family = family, data = working.df,
#> na.action = stats::na.omit)
#>
#> Coefficients:
#> (Intercept) bmicat1 bmicat2 bmicat3 AGE SEX1
#> -5.73445 0.22184 0.09288 0.48711 0.10305 -0.80522
#> DIABETES1 CURSMOKE1 PREVHYP1
#> 1.50504 0.59057 0.76174
#>
#> Degrees of Freedom: 4220 Total (i.e. Null); 4212 Residual
#> (19 observations deleted due to missingness)
#> Null Deviance: 5735
#> Residual Deviance: 4686 AIC: 4704
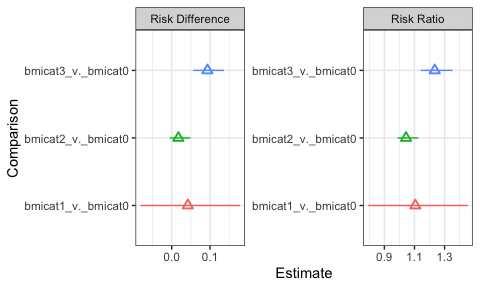
The results from the g-computation show the estimated risk difference and ratio, in addition to other information. From these results, we see that obese persons have an 11.6% (95% CI: 7.2, 16.6) increase in 24-year risk of cardiovascular disease or death compared to normal weight persons. Underweight persons also have increased risk, more so than overweight persons. Not surprisingly, the estimate comparing underweight to normal weight persons is imprecise given the few people in the dataset who were underweight.
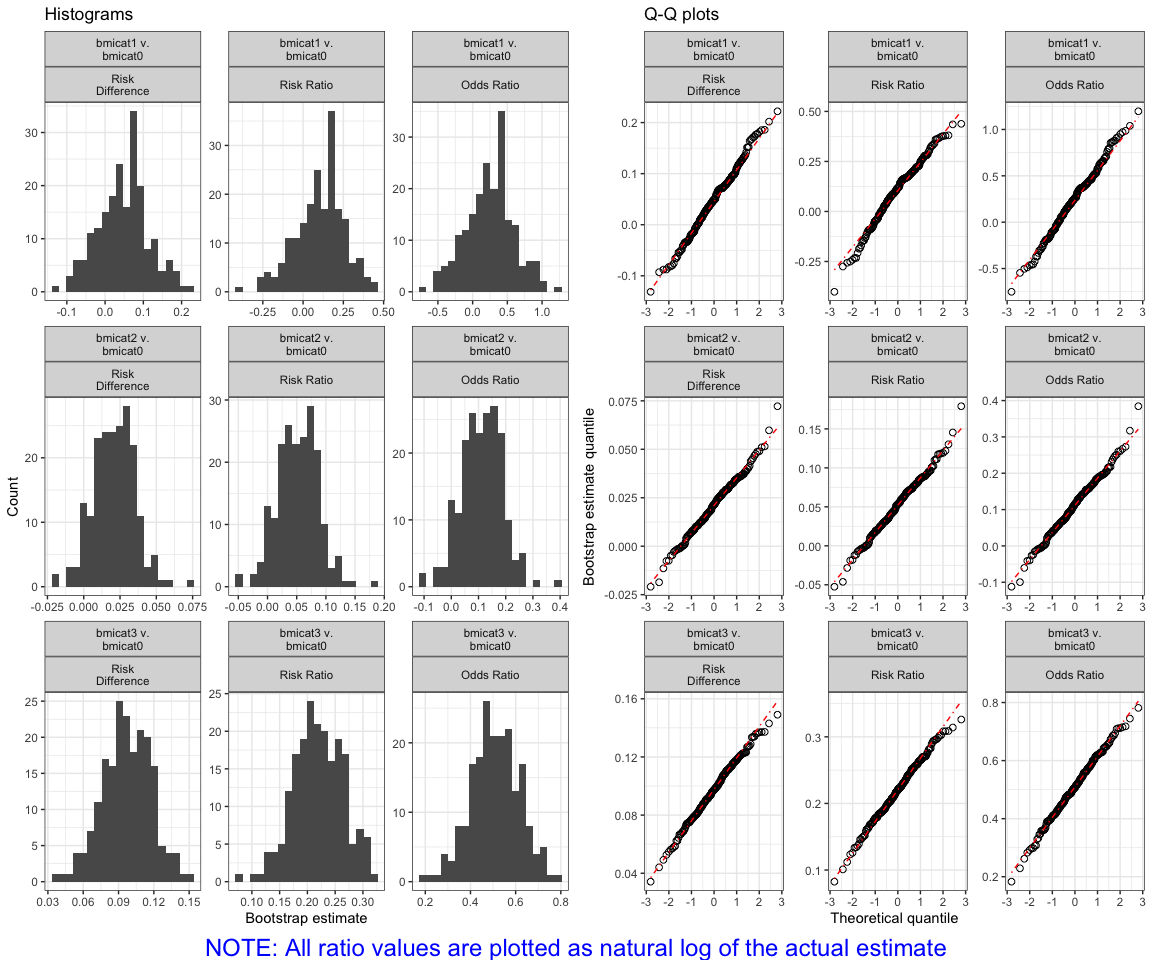
You can verify that the parameter estimates from the bootstraps are normally distributed:
plot(bmi.results)
#> Warning in log(as.numeric(.data$value)): NaNs produced
You can also easily plot the outcome estimates:
ggplot(bmi.results$results.df %>%
filter(Parameter %in% c("Risk Difference", "Risk Ratio"))
) +
geom_pointrange(aes(x = Comparison,
y = Estimate,
ymin = `2.5% CL`,
ymax = `97.5% CL`,
color = Comparison),
shape = 2
) +
coord_flip() +
facet_wrap(~Parameter, scale = "free") +
theme_bw() +
theme(legend.position = "none")