Visualize the Results of Risk-of-Bias (ROB) Assessments.
robvis 
UPDATE: robvis now exists as a web-app, aimed at those who are not familiar with R or who want to explore the package’s functionality before installing it locally.
Description
The robvis package takes the summary table from risk-of-bias assessments, converts it to tidy data, and produces summary plots formatted according to the assessment tool used.
Getting started
First ensure you have the devtools package installed:
install.packages("devtools")
library(devtools)
Then, to install:
install_github("mcguinlu/robvis")
library(robvis)
To update the package, run the install_github("mcguinlu/robvis") command again.
Load data
To load your own data from a .csv file:
mydata <- read.csv("path/to/mydata.csv", header = TRUE)
To help users explore robvis, we have included example datasets in the package, one for each of the tool templates that currently exist within the package. The data_rob2 dataset (view it here), which contains example risk-of-bias assessments performed using the RoB2.0 tool for randomized controlled trials, is used to create the plots in subsequent sections.
Create plots
The package contains two plotting functions:
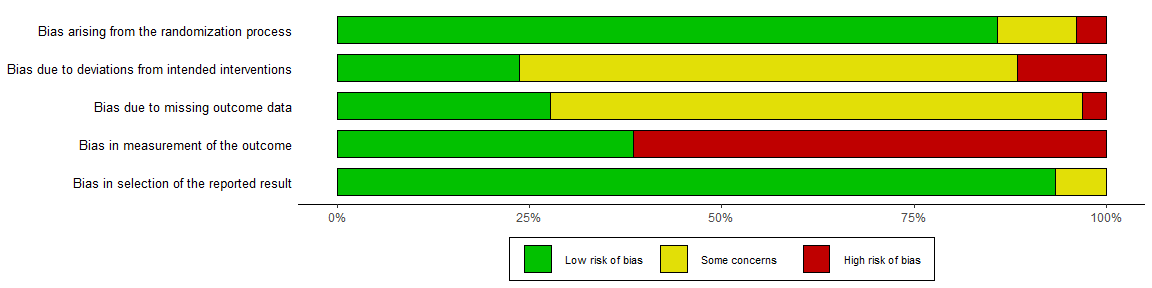
1. rob_summary()
Returns a ggplot object displaying a weighted barchart of the risk of bias of included studies across the domains of the specified tool.
summary_rob <- rob_summary(data = data_rob2, tool = "ROB2")
summary_rob
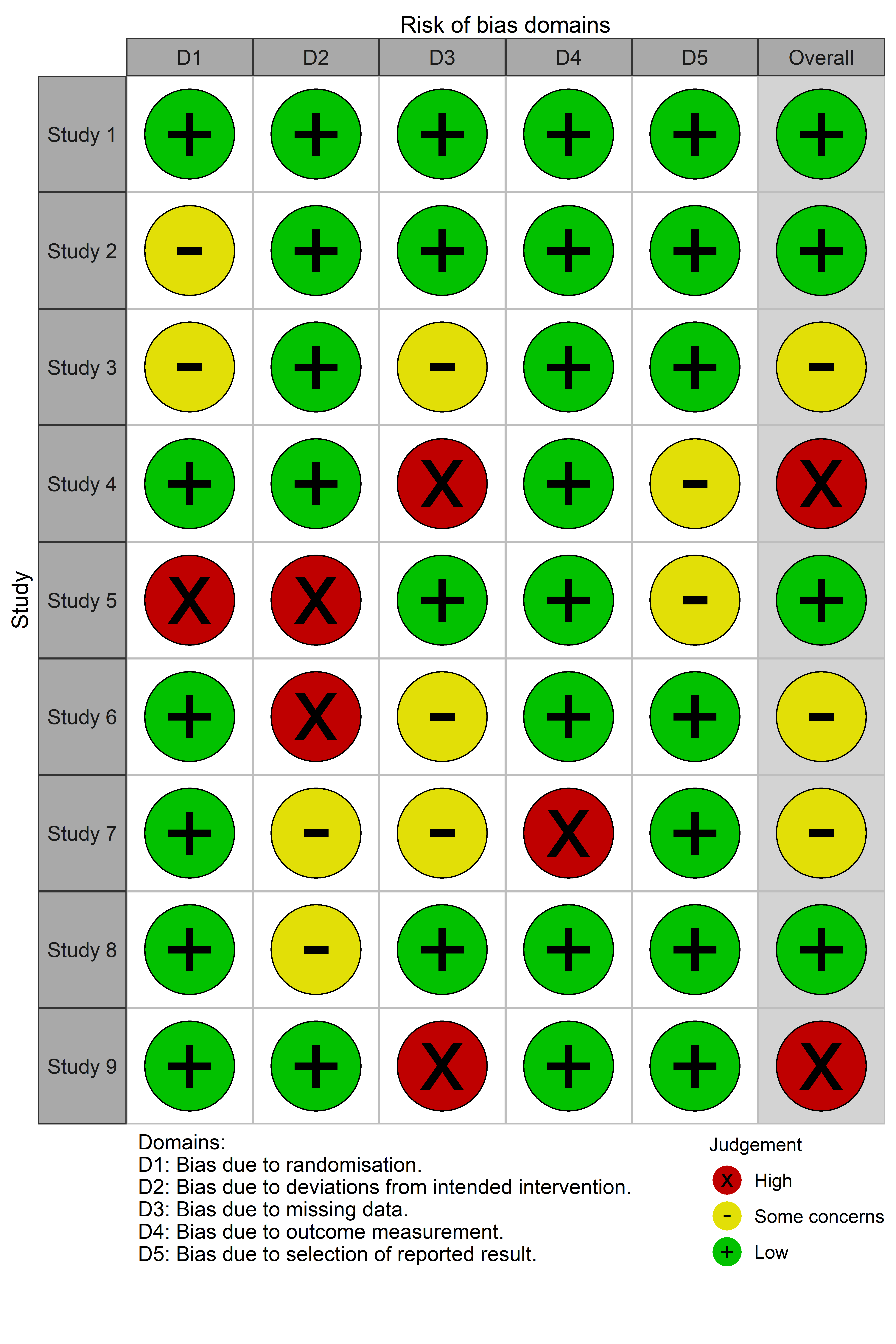
2. rob_traffic_light()
Returns a ggplot object displaying a “traffic light plot”, displaying the risk of bias judgment in each domain for each study.
trafficlight_rob <- rob_traffic_light(data = data_rob2, tool = "ROB2")
trafficlight_rob
Other functions
rob_tools()
Outputs a list of the risk of bias assessment tools for which a template currently exists in rob_summary(). We expect this list to be updated in the near future to include tools such as ROBIS (tool for assessing risk of bias in systematic reviews).
rob_tools()
[1] "ROB2"
[1] "ROBINS-I"
[1] "QUADAS-2"
[1] "ROB1"
Advanced usage
Change the colour scheme
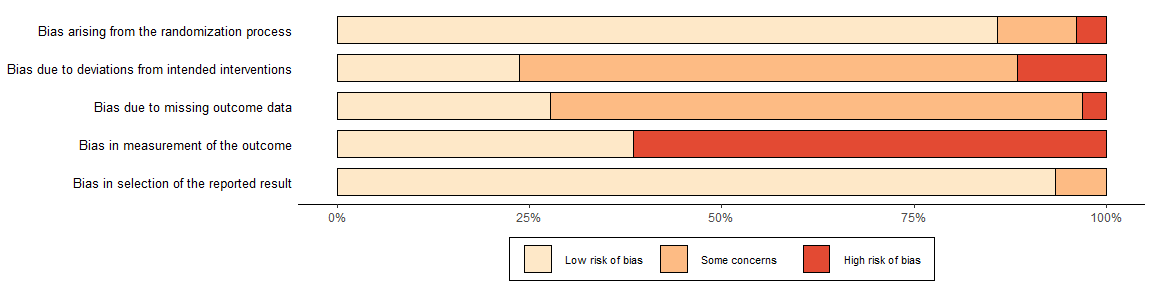
The colour argument of both plotting functions allows users to select from two predefined colour schemes (“cochrane” or “colourblind”) or to define their own palette by providing a vector of hex codes.
For example, to use the predefined “colourblind” palette:
summary_rob <- rob_summary(data = data_rob2, tool = "ROB2", colour = "colourblind")
summary_rob
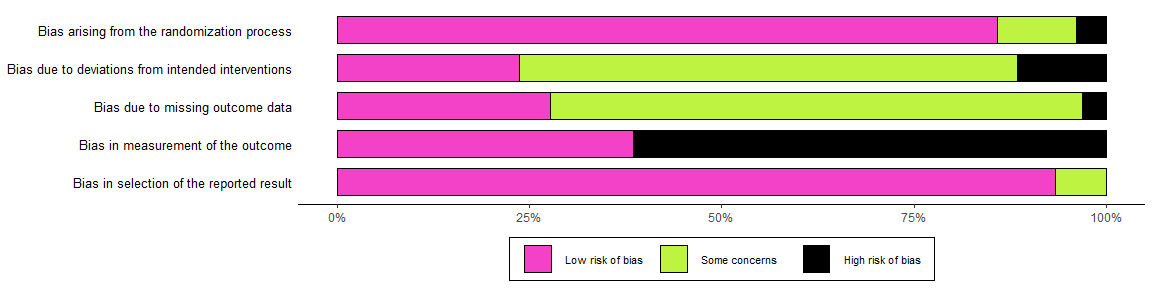
And to define your own colour scheme:
summary_rob <- rob_summary(data = data_rob2, tool = "ROB2", colour = c("#f442c8","#bef441","#000000"))
summary_rob
Created an unweighted summary barplot
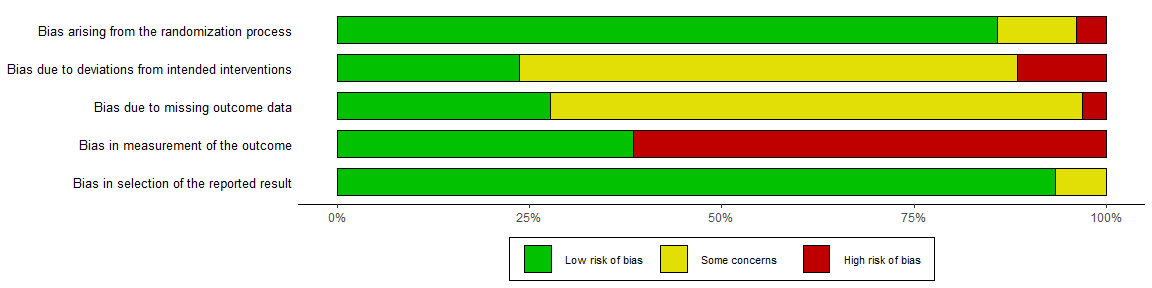
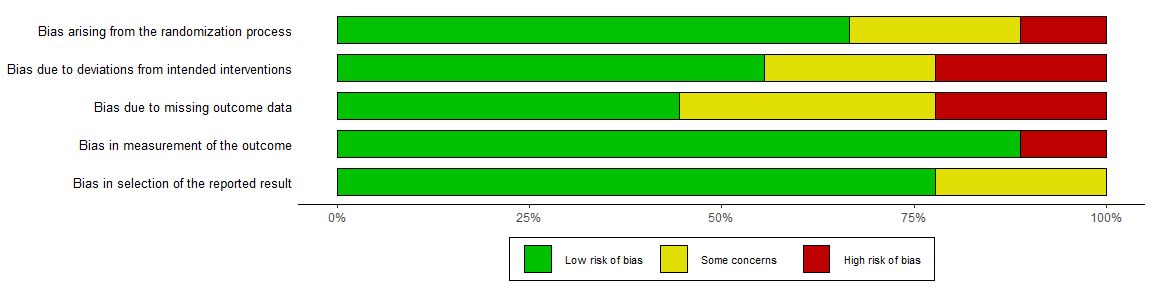
By default, the rob_summary() function creates a barplot weighted by some measure of a study’s precision. This can be prevented using the “weighted” argument. For example, compare the following two plots:
summary_rob <- rob_summary(data = data_rob2, tool = "ROB2")
summary_rob
summary_rob <- rob_summary(data = data_rob2, tool = "ROB2", weighted = FALSE)
summary_rob
Editing the plots
Finally, because the output (summary_rob and trafficlight_rob in the examples above) is a ggplot2 object, it is easy to adjust the plot to your own preferences.
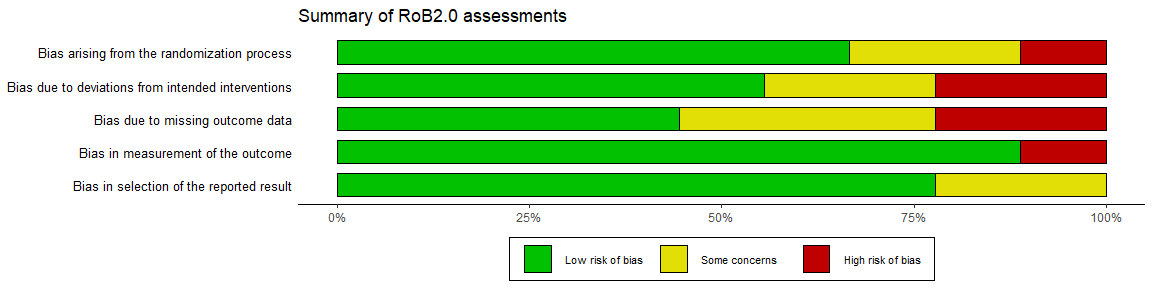
For example, to add a title to the unweighted RoB2.0 plot created above:
library(ggplot2)
summary_rob +
ggtitle("Summary of RoB2.0 assessments")
Code of conduct
Please note that the ‘robvis’ project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
License
This project is licensed under the MIT License - see the LICENSE.md file for details.
Acknowledgments
- The
rob_summary()function was based on code forwarded by a colleague. I recently discovered that this code was adapted from that presented in the wonderful “Doing Meta-Analysis in R” guide, so I would like to acknowledge the authors here. - Emily Kothe for help in fixing
ggplot2coding issues. - Eliza Grames for creating the
robvishex sticker.

