Get Silhouettes of Organisms from PhyloPic.
rphylopic 

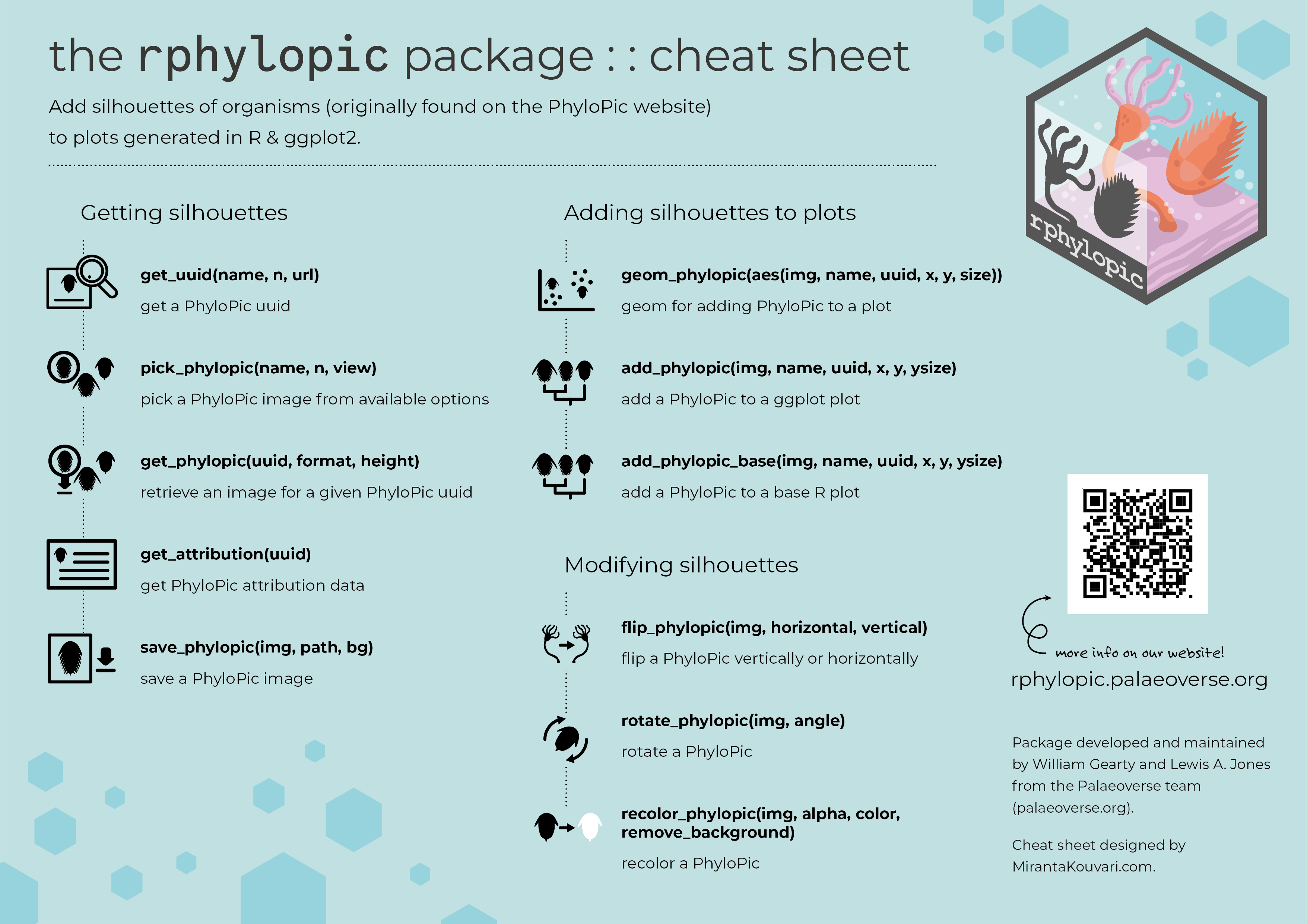
The purpose of the rphylopic package is to allow users to add silhouettes of organisms to plots generated in base R and ggplot2. To do so, it uses silhouettes made available via the PhyloPic website.
rphylopic was originally developed and maintained by Scott Chamberlain. From ver. 1.0.0, the package is now developed and maintained by William Gearty and Lewis A. Jones from the Palaeoverse team.
Installation
The stable version of rphylopic can be installed from CRAN using:
install.packages("rphylopic")
The development version of rphylopic can be installed via GitHub using:
install.packages("remotes")
remotes::install_github("palaeoverse/rphylopic")
How does it work?
Get an image uuid
# Load rphylopic
library(rphylopic)
# Get a single image uuid for a species
uuid <- get_uuid(name = "Canis lupus", n = 1)
# Get the image for that uuid
img <- get_phylopic(uuid = uuid)
# But multiple silhouettes can exist per species...
uuid <- get_uuid(name = "Canis lupus", n = 5)
Pick an image
# How do I pick?!
# It's difficult without seeing the image itself, let's use:
img <- pick_phylopic(name = "Canis lupus", n = 5)
Plot an image
Base R
# OK, now we've got the image we want... let's add it to a plot!
plot(x = 1, y = 1, type = "n")
add_phylopic_base(img = img, x = 1.25, y = 1.25, height = 0.25)
# But can't we just add an image straight away using the uuid? Sure!
uuid <- get_uuid(name = "Canis lupus", n = 1)
add_phylopic_base(uuid = uuid, x = 1, y = 1, height = 0.25)
# What about just using the first image linked to the name? Definitely!
add_phylopic_base(name = "Canis lupus", x = 0.75, y = 0.75, height = 0.25)
# Black is a bit boring? OK...
add_phylopic_base(name = "Canis lupus", x = 0.75, y = 1.25, height = 0.25, color = "orange")
ggplot2
# All of this functionality is available for ggplot2 as well...
# But we use add_phylopic and geom_phylopic instead!
library(ggplot2)
# Get image
uuid <- get_uuid(name = "Iris", n = 1)
img <- get_phylopic(uuid = uuid)
# Put a silhouette behind a plot
ggplot(iris) +
add_phylopic(x = 6.1, y = 3.2, img = img, alpha = 0.2) +
geom_point(aes(x = Sepal.Length, y = Sepal.Width))
# Plot silhouettes as points!
ggplot(iris) +
geom_phylopic(aes(x = Sepal.Length, y = Sepal.Width), img = img,
color = "purple", height = 0.25)
Get attribution
# PhyloPic has a lot of contributors and we should acknowledge
# their work. You can get data about images using get_attribution
# Get valid uuid
uuid <- get_uuid(name = "Nycticebus")
# Get attribution data for uuid
get_attribution(uuid = uuid)
Save an image
# How do I save an image?
# Get image
img <- pick_phylopic(name = "Phascolarctos cinereus", n = 1)
# Save image
save_phylopic(img = img)
How to contribute?
If you are interested in contributing to the rphylopic R package, you can do so by following these guidelines.
Code of Conduct
As with any community project, society, or meeting we feel it is important to established some expectations of behaviour. Please read our code of conduct, and reach out if you ever face any issues. Everyone has the right to live and work in a harassment-free environment.
Attribution
If you use the rphylopic package in your work, please acknowledge the contributors responsible for the image, acknowledge the creator of PhyloPic (Michael Keesey), and cite the following for rphylopic:
Gearty, W. and Jones, L.A. 2023. rphylopic: An R package for fetching, transforming, and visualising PhyloPic silhouettes. Methods in Ecology and Evolution, 14(11), 2700-2708. doi: 10.1111/2041-210X.14221.
Cheat sheet