Fitting Distributions to Given Data or Known Quantiles.
rriskDistributions
rriskDistributions is a collection of functions for fitting distributions to given data or known quantiles.
The two main functions fit.perc() and fit.cont() provide users a GUI that allows to choose a most appropriate distribution without any knowledge of the R syntax. Note that this package is part of the rrisk project.
E.g., we can fit random data generated from a gamma distribution with fit.cont():
res <- fit.cont(data2fit = rgamma(n = 37, shape = 4, rate = 0.08))
This will open a new window where the user can inspect diagnostic plots for a variety of possible distributions and then choose the distribution she wants to continue working with (the chosen distribution will be stored in the res variable):
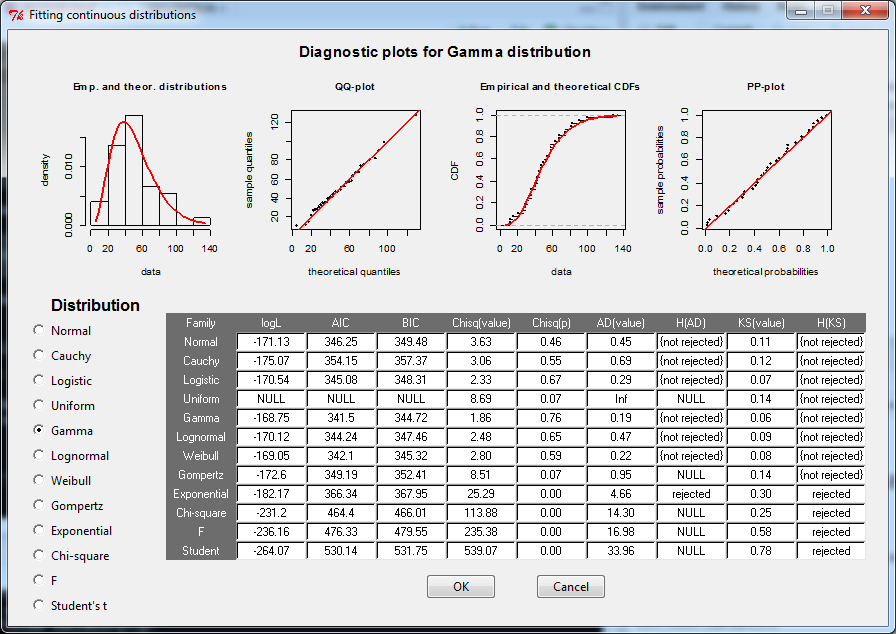
Updating to the latest version of rriskDistributions
You can track (and contribute to) development of rriskDistributions at https://github.com/BfRstats/rriskDistributions. To install it, run the following command (this requires the devtools package):
devtools::install_github("BfRstats/rriskDistributions")
Authors
- Natalia Belgorodski (STAT-UP Statistical Consulting)
- Matthias Greiner (Federal Institute for Risk Assessment, Germany)
- Kristin Tolksdorf (Federal Institute for Risk Assessment, Germany)
- Katharina Schueller (STAT-UP Statistical Consulting)
With contributions from
- Lutz Göhring (Lutz Göhring Consulting)
- Matthias Flor (Federal Institute for Risk Assessment, Germany)