Description
Learn Text 'Embeddings' with 'Starspace'.
Description
Wraps the 'StarSpace' library <https://github.com/facebookresearch/StarSpace> allowing users to calculate word, sentence, article, document, webpage, link and entity 'embeddings'. By using the 'embeddings', you can perform text based multi-label classification, find similarities between texts and categories, do collaborative-filtering based recommendation as well as content-based recommendation, find out relations between entities, calculate graph 'embeddings' as well as perform semi-supervised learning and multi-task learning on plain text. The techniques are explained in detail in the paper: 'StarSpace: Embed All The Things!' by Wu et al. (2017), available at <arXiv:1709.03856>.
README.md
ruimtehol: R package to Embed All the Things! using StarSpace
This repository contains an R package which wraps the StarSpace C++ library (https://github.com/facebookresearch/StarSpace), allowing the following:
- Text classification
- Learning word, sentence or document level embeddings
- Finding sentence or document similarity
- Ranking web documents
- Content-based recommendation (e.g. recommend text/music based on the content)
- Collaborative filtering based recommendation (e.g. recommend text/music based on interest)
- Identification of entity relationships
Installation
- For regular users, install the package from your local CRAN mirror
install.packages("ruimtehol") - For installing the development version of this package:
devtools::install_github("bnosac/ruimtehol", build_vignettes = TRUE)
Look to the vignette and the documentation of the functions
vignette("ground-control-to-ruimtehol", package = "ruimtehol")
help(package = "ruimtehol")
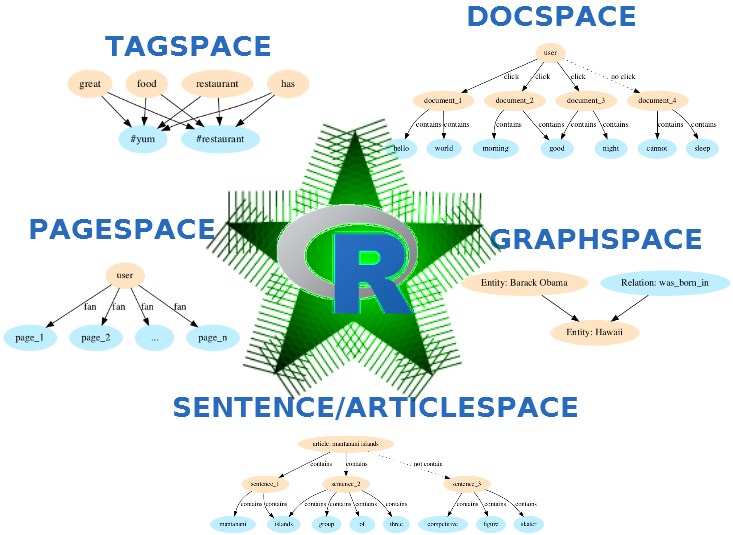
Main functionalities
This R package allows to Build Starspace models on your own text / Get embeddings of words/ngrams/sentences/documents/labels / Get predictions from a model (e.g. classification / ranking) / Get nearest neighbours similarity
The following functions are made available.
| Function | Functionality |
|---|---|
starspace | Low-level interface to build a Starspace model |
starspace_load_model | Load a pre-trained model or a tab-separated file |
starspace_save_model | Save a Starspace model |
starspace_embedding | Get embeddings of documents/words/ngrams/labels |
starspace_knn | Find k-nearest neighbouring information for new text |
starspace_dictonary | Get words/labels part of the model dictionary |
predict.textspace | Get predictions along a Starspace model |
as.matrix | Get words and label embeddings |
embedding_similarity | Cosine/dot product similarity between embeddings - top-n most similar text |
embed_wordspace | Build a Starspace model which calculates word/ngram embeddings |
embed_sentencespace | Build a Starspace model which calculates sentence embeddings |
embed_articlespace | Build a Starspace model for embedding articles - sentence-article similarities |
embed_tagspace | Build a Starspace model for multi-label classification |
embed_docspace | Build a Starspace model for content-based recommendation |
embed_pagespace | Build a Starspace model for interest-based recommendation |
embed_entityrelationspace | Build a Starspace model for entity relationship completion |
Example
Short example showing word embeddings
library(ruimtehol)
set.seed(123456789)
## Get some training data
download.file("https://s3.amazonaws.com/fair-data/starspace/wikipedia_train250k.tgz", "wikipedia_train250k.tgz")
x <- readLines("wikipedia_train250k.tgz", encoding = "UTF-8")
x <- x[-c(1:9)]
x <- x[sample(x = length(x), size = 10000)]
writeLines(text = x, sep = "\n", con = "wikipedia_train10k.txt")
## Train
set.seed(123456789)
model <- starspace(file = "wikipedia_train10k.txt", fileFormat = "labelDoc", dim = 10, trainMode = 3)
model
Object of class textspace
dimension of the embedding: 10
training arguments:
loss: hinge
margin: 0.05
similarity: cosine
epoch: 5
adagrad: TRUE
lr: 0.01
termLr: 1e-09
norm: 1
maxNegSamples: 10
negSearchLimit: 50
p: 0.5
shareEmb: TRUE
ws: 5
dropoutLHS: 0
dropoutRHS: 0
initRandSd: 0.001
embedding <- as.matrix(model)
embedding[c("school", "house"), ]
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
school 0.008395348 0.002858619 0.004770191 -0.03791502 -0.016193179 0.008368539 -0.0221493 0.01587386 -0.002012054 0.029385706
house 0.005371093 -0.007831781 0.010563998 0.01040361 0.000616577 0.005770847 -0.0097075 0.01678141 -0.004738560 0.009139475
dictionary <- starspace_dictionary(model)
## Save trained model as a binary file or as TSV so that you can inspect the embeddings e.g. with data.table::fread("wikipedia_embeddings.tsv")
starspace_save_model(model, file = "textspace.ruimtehol", method = "ruimtehol")
starspace_save_model(model, file = "wikipedia_embeddings.tsv", method = "tsv-data.table")
## Load a pre-trained model or pre-trained embeddings
model <- starspace_load_model("textspace.ruimtehol", method = "ruimtehol")
model <- starspace_load_model("wikipedia_embeddings.tsv", method = "tsv-data.table", trainMode = 3)
## Get the document embedding
starspace_embedding(model, "get the embedding of a full document")
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
get the embedding of a full document 0.1489144 -0.09543591 0.1242385 -0.1080941 0.6971645 0.3131362 -0.3405705 0.3293449 0.231894 -0.281555
The following functionalities do similar things. They see what is the closest word or sentence to a provided sentence.
## What is closest term from the dictionary
starspace_knn(model, "What does this bunch of text look like", k = 10)
## What is closest sentence to vector of sentences
predict(model, newdata = "what does this bunch of text look like",
basedoc = c("what does this bunch of text look like",
"word abracadabra was not part of the dictionary",
"give me back my mojo",
"cosine distance is what i show"))
## Get cosine distance between 2 sentence vectors
embedding_similarity(
starspace_embedding(model, "what does this bunch of text look like"),
starspace_embedding(model, "word abracadabra was not part of the dictionary"),
type = "cosine")
Short example showing classification modelling (tagspace)
Below Starspace is used for classification
library(ruimtehol)
data("dekamer", package = "ruimtehol")
dekamer$x <- strsplit(dekamer$question, "\\W")
dekamer$x <- sapply(dekamer$x, FUN = function(x) paste(setdiff(x, ""), collapse = " "))
dekamer$x <- tolower(dekamer$x)
dekamer$y <- strsplit(dekamer$question_theme, split = ",")
dekamer$y <- lapply(dekamer$y, FUN=function(x) gsub(" ", "-", x))
set.seed(123456789)
model <- embed_tagspace(x = dekamer$x, y = dekamer$y,
dim = 50,
lr = 0.01, epoch = 40, loss = "softmax", adagrad = TRUE,
similarity = "cosine", negSearchLimit = 50,
ngrams = 2, minCount = 2)
plot(model)
text <- c("de nmbs heeft het treinaanbod uitgebreid via onteigening ...",
"de migranten komen naar europa de asielcentra ...")
predict(model, text, k = 3)
predict(model, "koning filip", k = 10, type = "knn")
predict(model, "koning filip", k = 10, type = "embedding")
Notes
- Why did you call the package ruimtehol? Because that is the translation of StarSpace in WestVlaams.
- The R wrapper is distributed under the Mozilla Public License 2.0. The package contains a copy of the StarSpace C++ code (namely all code under src/Starspace) which has a BSD license (which is available in file LICENSE.notes) and also has an accompanying PATENTS file which you can inspect here.
Support in text mining
Need support in text mining? Contact BNOSAC: http://www.bnosac.be.