Real-Time PCR Data Sets by Rutledge et al. (2004).
rutledge
{rutledge} is an R data package that provides real-time PCR raw fluorescence data by Rutledge et al. (2004) in tidy format.
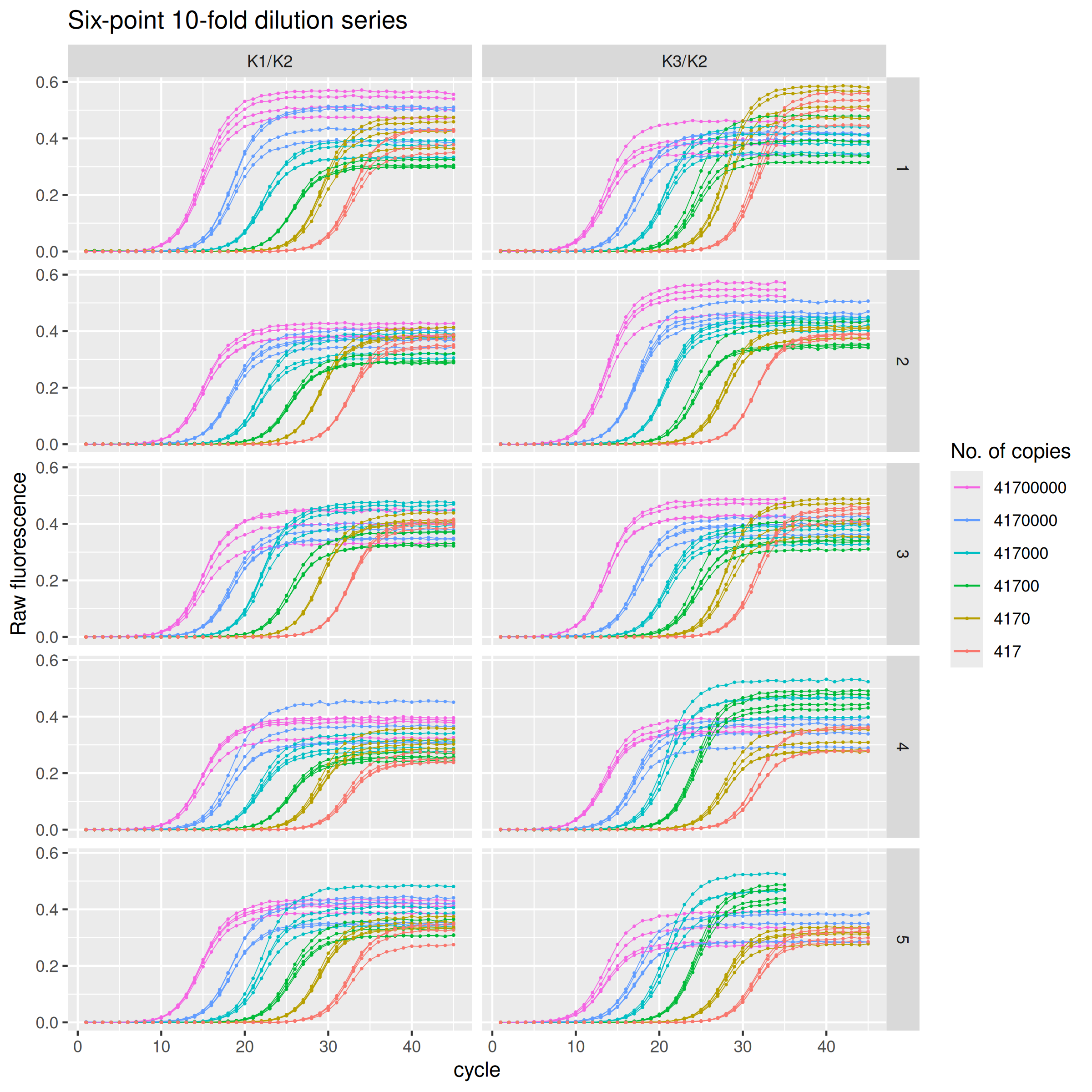
The eponymous data set rutledge comprises a six-point, ten-fold dilution series repeated in 5 independent runs. In each run, for each concentration there are four replicates. Of a total of 240 amplification curves, 212 curves are 45 cycles long and 28 curves are 35 cycles long only. The data is for two targets (amplicons): K1/K2, 102 bp, and K3/K2, 218 bp.
Installation
Install {rutledge} from CRAN:
# Install from CRAN
install.packages("rutledge")
You can install the development version of {rutledge} like so:
# install.packages("remotes")
remotes::install_github("ramiromagno/rutledge")
Data
rutledge is provided as a tidy data set, in long format, i.e. each row is for an amplication curve point (cycle/fluor).
library(rutledge)
rutledge
#> # A tibble: 10,800 × 10
#> plate well dye target sample_type replicate copies dilution cycle fluor
#> <fct> <fct> <chr> <fct> <fct> <int> <int> <int> <dbl> <dbl>
#> 1 1 <NA> SYBR K1/K2 std 1 41700000 1 1 0
#> 2 1 <NA> SYBR K1/K2 std 1 41700000 1 2 0
#> 3 1 <NA> SYBR K1/K2 std 1 41700000 1 3 0
#> 4 1 <NA> SYBR K1/K2 std 1 41700000 1 4 0
#> 5 1 <NA> SYBR K1/K2 std 1 41700000 1 5 0.0007
#> 6 1 <NA> SYBR K1/K2 std 1 41700000 1 6 0.0022
#> 7 1 <NA> SYBR K1/K2 std 1 41700000 1 7 0.0005
#> 8 1 <NA> SYBR K1/K2 std 1 41700000 1 8 0.0047
#> 9 1 <NA> SYBR K1/K2 std 1 41700000 1 9 0.0107
#> 10 1 <NA> SYBR K1/K2 std 1 41700000 1 10 0.0203
#> # ℹ 10,790 more rows
The rutledge data set comprises 240 amplification curves: 2 amplicons $\times$ 5 runs (plates) $\times$ 6 dilution levels $\times$ 4 replicates.
rutledge |>
dplyr::count(plate, target, copies, replicate)
#> # A tibble: 240 × 5
#> plate target copies replicate n
#> <fct> <fct> <int> <int> <int>
#> 1 1 K1/K2 417 1 45
#> 2 1 K1/K2 417 2 45
#> 3 1 K1/K2 417 3 45
#> 4 1 K1/K2 417 4 45
#> 5 1 K1/K2 4170 1 45
#> 6 1 K1/K2 4170 2 45
#> 7 1 K1/K2 4170 3 45
#> 8 1 K1/K2 4170 4 45
#> 9 1 K1/K2 41700 1 45
#> 10 1 K1/K2 41700 2 45
#> # ℹ 230 more rows
rutledge |>
ggplot(mapping = aes(
x = cycle,
y = fluor,
group = interaction(plate, target, copies, replicate),
col = as.character(copies)
)) +
geom_line(linewidth = 0.2) +
geom_point(size = 0.2) +
labs(y = "Raw fluorescence", colour = "No. of copies", title = "Six-point 10-fold dilution series") +
guides(color = guide_legend(override.aes = list(linewidth = 0.5), reverse = TRUE)) +
facet_grid(rows = vars(plate), cols = vars(target))
#> Warning: Removed 280 rows containing missing values or values outside the scale range
#> (`geom_line()`).
#> Warning: Removed 280 rows containing missing values or values outside the scale range
#> (`geom_point()`).
Code of Conduct
Please note that the rutledge project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
References
R. G. Rutledge. Sigmoidal curve-fitting redefines quantitative real-time PCR with the prospective of developing automated high-throughput applications. Nucleic Acids Research 32:e178 (2004). doi: 10.1093/nar/gnh177.