Spatial Association Between Regionalizations.
sabre 
The sabre (Spatial Association Between REgionalizations) is an R package for calculating a degree of spatial association between regionalizations or categorical maps. This package offers support for sf, RasterLayer, SpatRaster, and stars spatial objects, and the following methods:
- the V-measure method (Nowosad and Stepinski, 2018)
- the MapCurve method (Hargrove et al., 2006)
Installation
You can install the released version of sabre from CRAN with:
install.packages("sabre")
You can install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("Nowosad/sabre")
Example
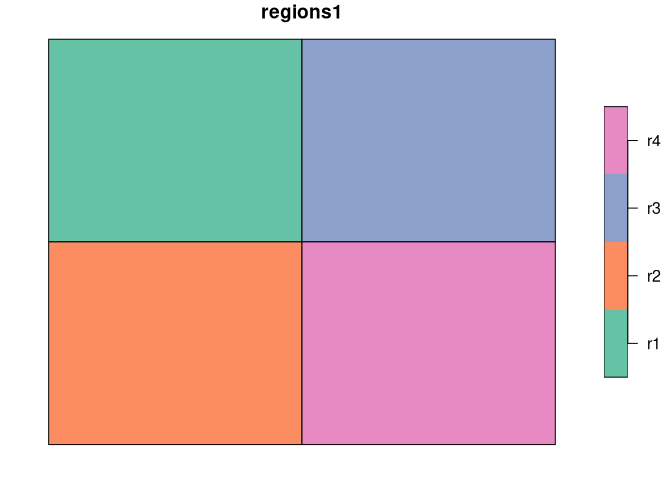
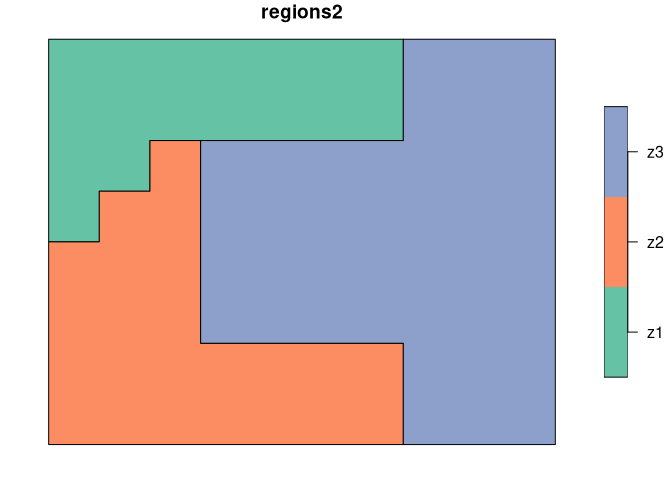
We use two simple regionalization, regions1 and regions2 to show the basic concept of calculating a degree of spatial association.
library(sabre)
library(sf)
data("regions1")
data("regions2")
The first map, regions1 consists of four regions of the same shape and size, while the second one, regions2 has three irregular regions.
The vmeasure_calc() function allows for calculation of a degree of spatial association between regionalizations or categorical maps using the information-theoretical V-measure. It requires, at least, four arguments:
x- ansfobject containing the first regionalizationx_name- a name of the column with regions names of the first regionalizationy- ansfobject containing the second regionalizationy_name- a name of the column with regions names of the second regionalization
regions_vm = vmeasure_calc(x = regions1, y = regions2, x_name = z, y_name = z)
The result is a list with three metrics of spatial association - V-measure, Homogeneity, Completeness - and two sf objects with preprocessed input maps - $map1 and $map2.
regions_vm
#> The SABRE results:
#>
#> V-measure: 0.36
#> Homogeneity: 0.32
#> Completeness: 0.42
#>
#> The spatial objects can be retrieved with:
#> $map1 - the first map
#> $map2 - the second map
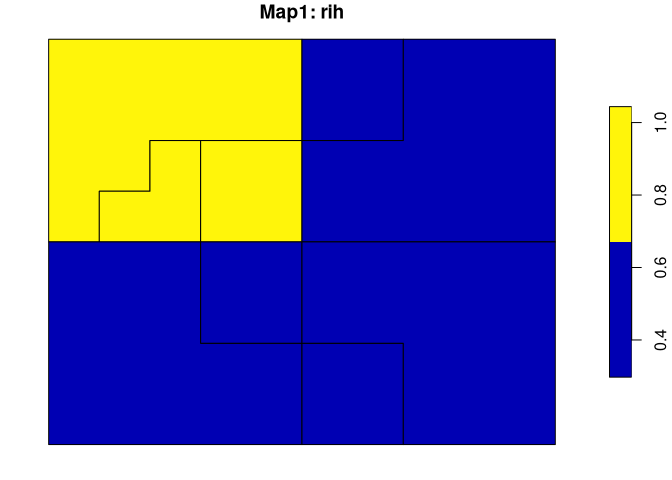
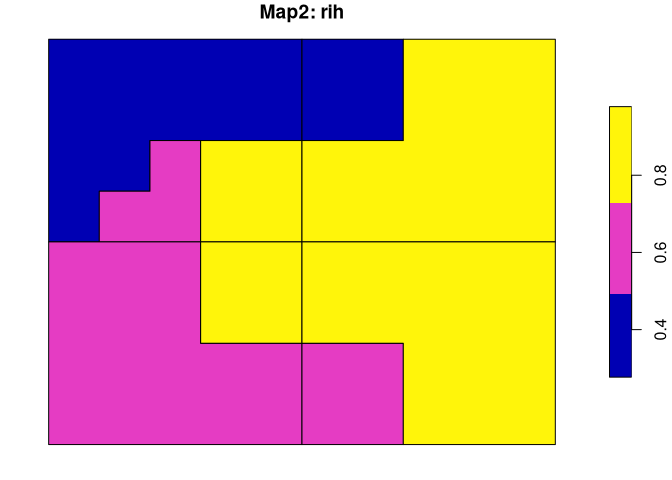
Both spatial outputs have two columns. The first one contains regions’ names/values and the second one (rih) describes regions’ inhomogeneities.
plot(regions_vm$map1["rih"], main = "Map1: rih")
plot(regions_vm$map2["rih"], main = "Map2: rih")
More examples can be found in the package vignette and in the sabre: or how to compare two maps? blog post.
Logo
Hex logo was created with hexmake using icons made by Smashicons and Creaticca Creative Agency from www.flaticon.com.
References
- Nowosad, Jakub, and Tomasz F. Stepinski. “Spatial association between regionalizations using the information-theoretical V-measure.” International Journal of Geographical Information Science (2018). https://doi.org/10.1080/13658816.2018.1511794
- Rosenberg, Andrew, and Julia Hirschberg. “V-measure: A conditional entropy-based external cluster evaluation measure.” Proceedings of the 2007 joint conference on empirical methods in natural language processing and computational natural language learning (EMNLP-CoNLL). 2007.
- Hargrove, William W., Forrest M. Hoffman, and Paul F. Hessburg. “Mapcurves: a quantitative method for comparing categorical maps.” Journal of Geographical Systems 8.2 (2006): 187.