Statistical Models for the Unsupervised Segmentation of Time-Series ('SaMUraiS').
SaMUraiS: StAtistical Models for the UnsupeRvised segmentAtIon of time-Series
samurais is an open source toolbox (available in R and in Matlab) including many original and flexible user-friendly statistical latent variable models and unsupervised algorithms to segment and represent, time-series data (univariate or multivariate), and more generally, longitudinal data which include regime changes.
Our samurais use mainly the following efficient “sword” packages to segment data: Regression with Hidden Logistic Process (RHLP), Hidden Markov Model Regression (HMMR), Piece-Wise regression (PWR), Multivariate ‘RHLP’ (MRHLP), and Multivariate ‘HMMR’ (MHMMR).
The models and algorithms are developed and written in Matlab by Faicel Chamroukhi, and translated and designed into R packages by Florian Lecocq, Marius Bartcus and Faicel Chamroukhi.
Installation
You can install the samurais package from GitHub with:
# install.packages("devtools")
devtools::install_github("fchamroukhi/SaMUraiS")
To build vignettes for examples of usage, type the command below instead:
# install.packages("devtools")
devtools::install_github("fchamroukhi/SaMUraiS",
build_opts = c("--no-resave-data", "--no-manual"),
build_vignettes = TRUE)
Use the following command to display vignettes:
browseVignettes("samurais")
Usage
library(samurais)
RHLP
# Application to a toy data set
data("univtoydataset")
K <- 5 # Number of regimes (mixture components)
p <- 3 # Dimension of beta (order of the polynomial regressors)
q <- 1 # Dimension of w (order of the logistic regression: to be set to 1 for segmentation)
variance_type <- "heteroskedastic" # "heteroskedastic" or "homoskedastic" model
n_tries <- 1
max_iter = 1500
threshold <- 1e-6
verbose <- TRUE
verbose_IRLS <- FALSE
rhlp <- emRHLP(univtoydataset$x, univtoydataset$y, K, p, q,
variance_type, n_tries, max_iter, threshold,
verbose, verbose_IRLS)
#> EM: Iteration : 1 || log-likelihood : -2119.27308534609
#> EM: Iteration : 2 || log-likelihood : -1149.01040321999
#> EM: Iteration : 3 || log-likelihood : -1118.20384281234
#> EM: Iteration : 4 || log-likelihood : -1096.88260636121
#> EM: Iteration : 5 || log-likelihood : -1067.55719357295
#> EM: Iteration : 6 || log-likelihood : -1037.26620122646
#> EM: Iteration : 7 || log-likelihood : -1022.71743069484
#> EM: Iteration : 8 || log-likelihood : -1006.11825447077
#> EM: Iteration : 9 || log-likelihood : -1001.18491883952
#> EM: Iteration : 10 || log-likelihood : -1000.91250763556
#> EM: Iteration : 11 || log-likelihood : -1000.62280600209
#> EM: Iteration : 12 || log-likelihood : -1000.3030988811
#> EM: Iteration : 13 || log-likelihood : -999.932334880131
#> EM: Iteration : 14 || log-likelihood : -999.484219706691
#> EM: Iteration : 15 || log-likelihood : -998.928118038989
#> EM: Iteration : 16 || log-likelihood : -998.234244664472
#> EM: Iteration : 17 || log-likelihood : -997.359536276056
#> EM: Iteration : 18 || log-likelihood : -996.152654857298
#> EM: Iteration : 19 || log-likelihood : -994.697863447307
#> EM: Iteration : 20 || log-likelihood : -993.186583974542
#> EM: Iteration : 21 || log-likelihood : -991.81352379631
#> EM: Iteration : 22 || log-likelihood : -990.611295217008
#> EM: Iteration : 23 || log-likelihood : -989.539226273251
#> EM: Iteration : 24 || log-likelihood : -988.55311887915
#> EM: Iteration : 25 || log-likelihood : -987.539963690533
#> EM: Iteration : 26 || log-likelihood : -986.073920116541
#> EM: Iteration : 27 || log-likelihood : -983.263549878169
#> EM: Iteration : 28 || log-likelihood : -979.340492188909
#> EM: Iteration : 29 || log-likelihood : -977.468559852711
#> EM: Iteration : 30 || log-likelihood : -976.653534236095
#> EM: Iteration : 31 || log-likelihood : -976.5893387433
#> EM: Iteration : 32 || log-likelihood : -976.589338067237
rhlp$summary()
#> ---------------------
#> Fitted RHLP model
#> ---------------------
#>
#> RHLP model with K = 5 components:
#>
#> log-likelihood nu AIC BIC ICL
#> -976.5893 33 -1009.589 -1083.959 -1083.176
#>
#> Clustering table (Number of observations in each regimes):
#>
#> 1 2 3 4 5
#> 100 120 200 100 150
#>
#> Regression coefficients:
#>
#> Beta(K = 1) Beta(K = 2) Beta(K = 3) Beta(K = 4) Beta(K = 5)
#> 1 6.031875e-02 -5.434903 -2.770416 120.7699 4.027542
#> X^1 -7.424718e+00 158.705091 43.879453 -474.5888 13.194261
#> X^2 2.931652e+02 -650.592347 -94.194780 597.7948 -33.760603
#> X^3 -1.823560e+03 865.329795 67.197059 -244.2386 20.402153
#>
#> Variances:
#>
#> Sigma2(K = 1) Sigma2(K = 2) Sigma2(K = 3) Sigma2(K = 4) Sigma2(K = 5)
#> 1.220624 1.110243 1.079394 0.9779734 1.028332
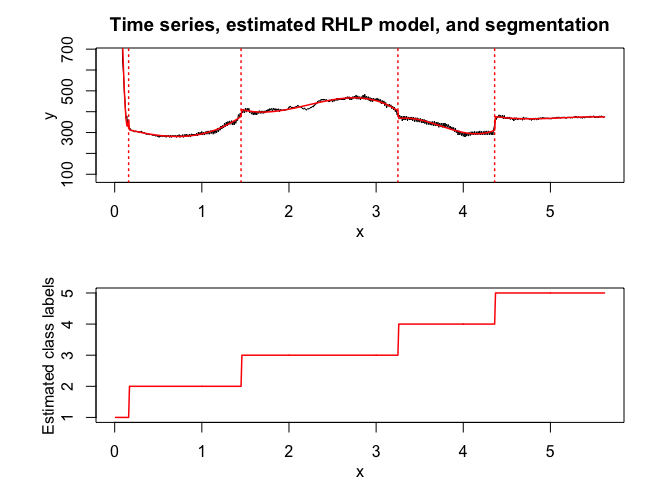
rhlp$plot()
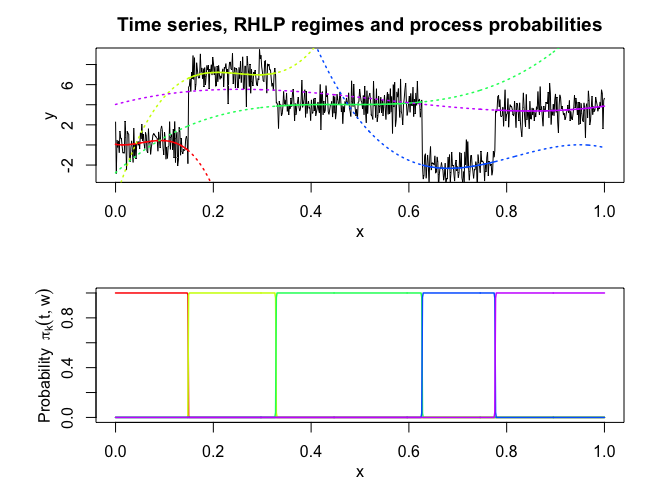
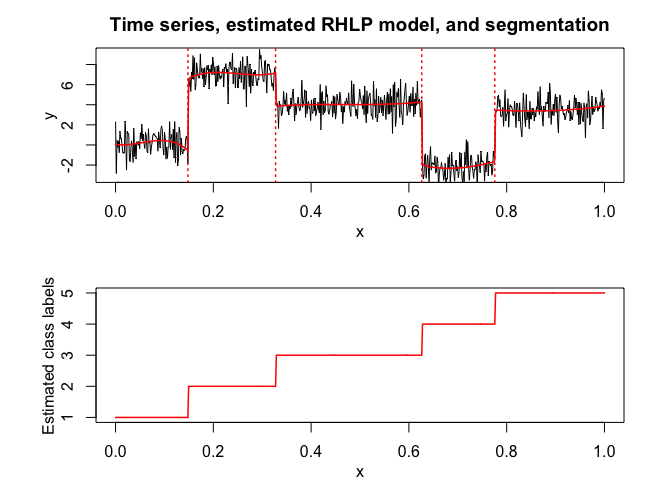
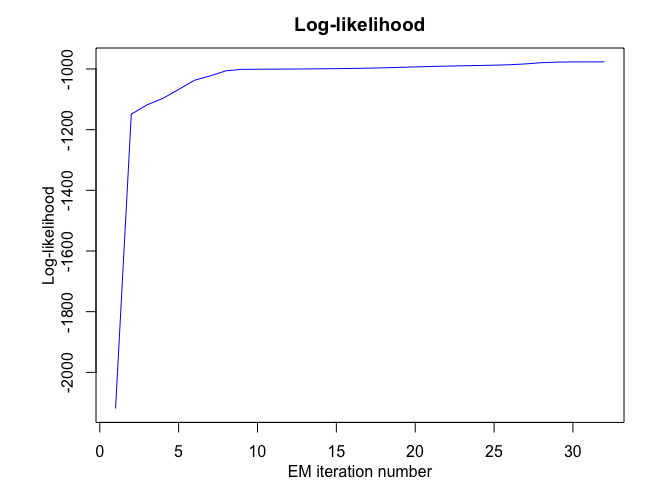
# Application to a real data set
data("univrealdataset")
K <- 5 # Number of regimes (mixture components)
p <- 3 # Dimension of beta (order of the polynomial regressors)
q <- 1 # Dimension of w (order of the logistic regression: to be set to 1 for segmentation)
variance_type <- "heteroskedastic" # "heteroskedastic" or "homoskedastic" model
n_tries <- 1
max_iter = 1500
threshold <- 1e-6
verbose <- FALSE
verbose_IRLS <- FALSE
rhlp <- emRHLP(univrealdataset$x, univrealdataset$y2, K, p, q,
variance_type, n_tries, max_iter, threshold,
verbose, verbose_IRLS)
rhlp$summary()
#> ---------------------
#> Fitted RHLP model
#> ---------------------
#>
#> RHLP model with K = 5 components:
#>
#> log-likelihood nu AIC BIC ICL
#> -1946.213 33 -1979.213 -2050.683 -2050.449
#>
#> Clustering table (Number of observations in each regimes):
#>
#> 1 2 3 4 5
#> 16 129 180 111 126
#>
#> Regression coefficients:
#>
#> Beta(K = 1) Beta(K = 2) Beta(K = 3) Beta(K = 4) Beta(K = 5)
#> 1 2187.539 330.05723 1508.2809 -13446.7332 6417.62830
#> X^1 -15032.659 -107.79782 -1648.9562 11321.4509 -3571.94090
#> X^2 -56433.432 14.40154 786.5723 -3062.2825 699.55894
#> X^3 494014.670 56.88016 -118.0693 272.7844 -45.42922
#>
#> Variances:
#>
#> Sigma2(K = 1) Sigma2(K = 2) Sigma2(K = 3) Sigma2(K = 4) Sigma2(K = 5)
#> 8924.363 49.22616 78.2758 105.6606 15.66317
rhlp$plot()
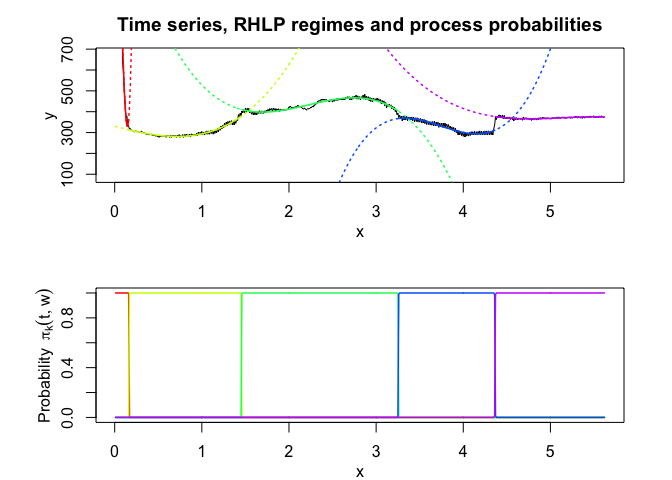
HMMR
# Application to a toy data set
data("univtoydataset")
K <- 5 # Number of regimes (states)
p <- 3 # Dimension of beta (order of the polynomial regressors)
variance_type <- "heteroskedastic" # "heteroskedastic" or "homoskedastic" model
n_tries <- 1
max_iter <- 1500
threshold <- 1e-6
verbose <- TRUE
hmmr <- emHMMR(univtoydataset$x, univtoydataset$y, K, p, variance_type,
n_tries, max_iter, threshold, verbose)
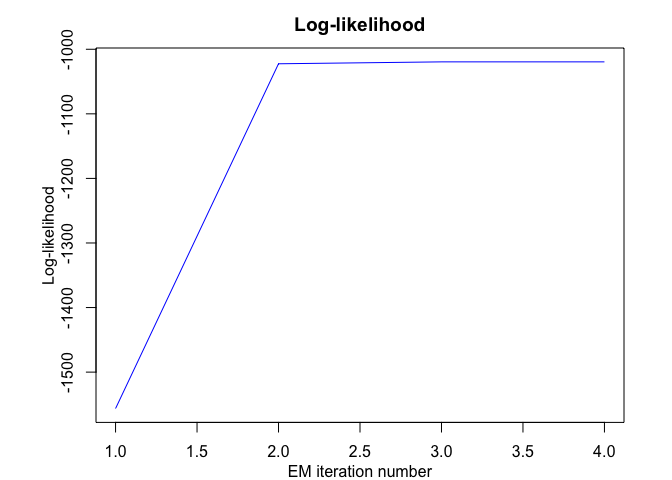
#> EM: Iteration : 1 || log-likelihood : -1556.39696825601
#> EM: Iteration : 2 || log-likelihood : -1022.47935723687
#> EM: Iteration : 3 || log-likelihood : -1019.51830707432
#> EM: Iteration : 4 || log-likelihood : -1019.51780361388
hmmr$summary()
#> ---------------------
#> Fitted HMMR model
#> ---------------------
#>
#> HMMR model with K = 5 components:
#>
#> log-likelihood nu AIC BIC
#> -1019.518 49 -1068.518 -1178.946
#>
#> Clustering table (Number of observations in each regimes):
#>
#> 1 2 3 4 5
#> 100 120 200 100 150
#>
#> Regression coefficients:
#>
#> Beta(K = 1) Beta(K = 2) Beta(K = 3) Beta(K = 4) Beta(K = 5)
#> 1 6.031872e-02 -5.326689 -2.65064 120.8612 3.858683
#> X^1 -7.424715e+00 157.189455 43.13601 -474.9870 13.757279
#> X^2 2.931651e+02 -643.706204 -92.68115 598.3726 -34.384734
#> X^3 -1.823559e+03 855.171715 66.18499 -244.5175 20.632196
#>
#> Variances:
#>
#> Sigma2(K = 1) Sigma2(K = 2) Sigma2(K = 3) Sigma2(K = 4) Sigma2(K = 5)
#> 1.220624 1.111487 1.080043 0.9779724 1.028399
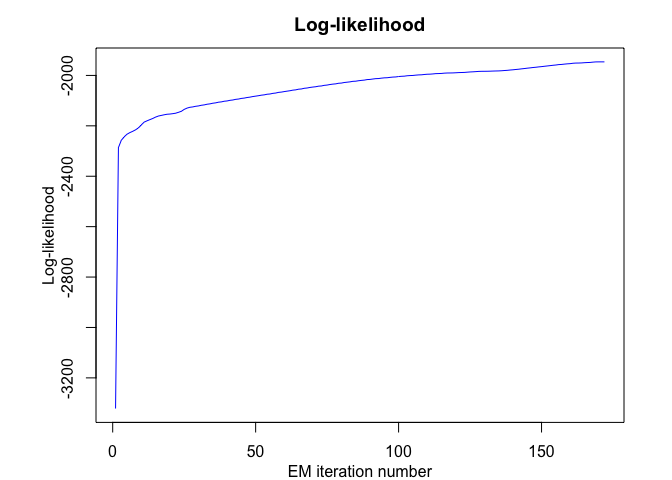
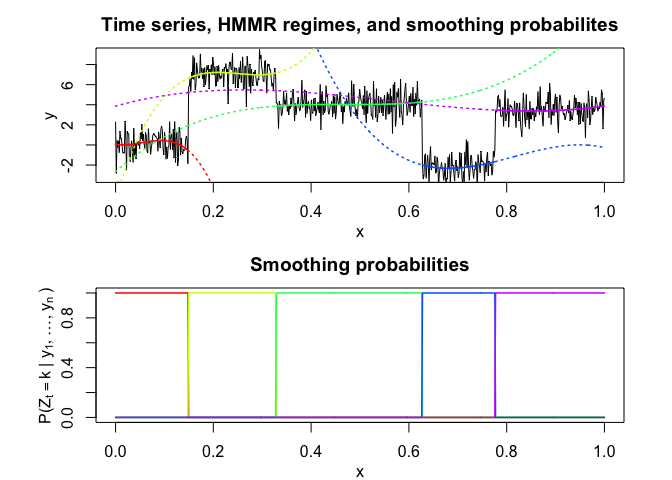
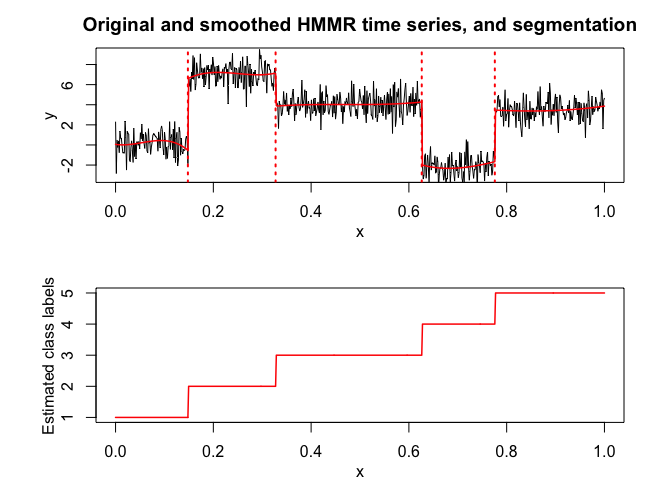
hmmr$plot(what = c("smoothed", "regressors", "loglikelihood"))
# Application to a real data set
data("univrealdataset")
K <- 5 # Number of regimes (states)
p <- 3 # Dimension of beta (order of the polynomial regressors)
variance_type <- "heteroskedastic" # "heteroskedastic" or "homoskedastic" model
n_tries <- 1
max_iter <- 1500
threshold <- 1e-6
verbose <- TRUE
hmmr <- emHMMR(univrealdataset$x, univrealdataset$y2, K, p, variance_type,
n_tries, max_iter, threshold, verbose)
#> EM: Iteration : 1 || log-likelihood : -2733.41028643114
#> EM: Iteration : 2 || log-likelihood : -2303.24018378559
#> EM: Iteration : 3 || log-likelihood : -2295.0470677529
#> EM: Iteration : 4 || log-likelihood : -2288.57866215726
#> EM: Iteration : 5 || log-likelihood : -2281.36756202518
#> EM: Iteration : 6 || log-likelihood : -2273.50303676091
#> EM: Iteration : 7 || log-likelihood : -2261.70334656117
#> EM: Iteration : 8 || log-likelihood : -2243.43509121433
#> EM: Iteration : 9 || log-likelihood : -2116.4610801575
#> EM: Iteration : 10 || log-likelihood : -2046.73194777839
#> EM: Iteration : 11 || log-likelihood : -2046.68328282973
#> EM: Iteration : 12 || log-likelihood : -2046.67329222076
#> EM: Iteration : 13 || log-likelihood : -2046.66915144265
#> EM: Iteration : 14 || log-likelihood : -2046.66694236131
#> EM: Iteration : 15 || log-likelihood : -2046.66563379017
hmmr$summary()
#> ---------------------
#> Fitted HMMR model
#> ---------------------
#>
#> HMMR model with K = 5 components:
#>
#> log-likelihood nu AIC BIC
#> -2046.666 49 -2095.666 -2201.787
#>
#> Clustering table (Number of observations in each regimes):
#>
#> 1 2 3 4 5
#> 14 214 99 109 126
#>
#> Regression coefficients:
#>
#> Beta(K = 1) Beta(K = 2) Beta(K = 3) Beta(K = 4) Beta(K = 5)
#> 1 2152.64 379.75158 5211.1759 -14306.4654 6417.62823
#> X^1 -12358.67 -373.37266 -5744.7879 11987.6666 -3571.94086
#> X^2 -103908.33 394.49359 2288.9418 -3233.8021 699.55894
#> X^3 722173.26 -98.60485 -300.7686 287.4567 -45.42922
#>
#> Variances:
#>
#> Sigma2(K = 1) Sigma2(K = 2) Sigma2(K = 3) Sigma2(K = 4) Sigma2(K = 5)
#> 9828.793 125.3346 58.71053 105.8328 15.66317
hmmr$plot(what = c("smoothed", "regressors", "loglikelihood"))
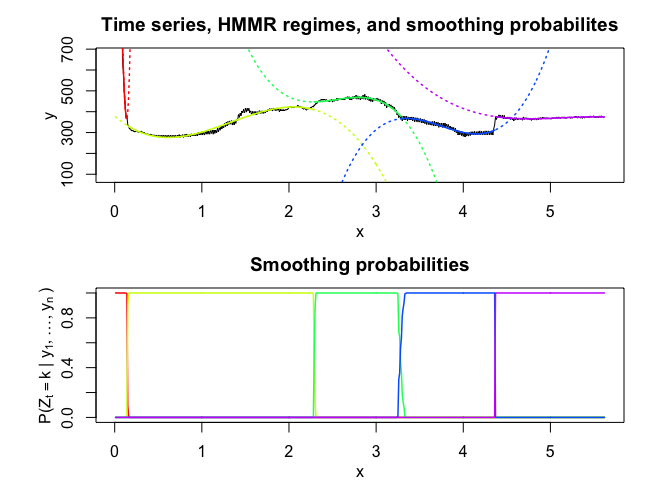
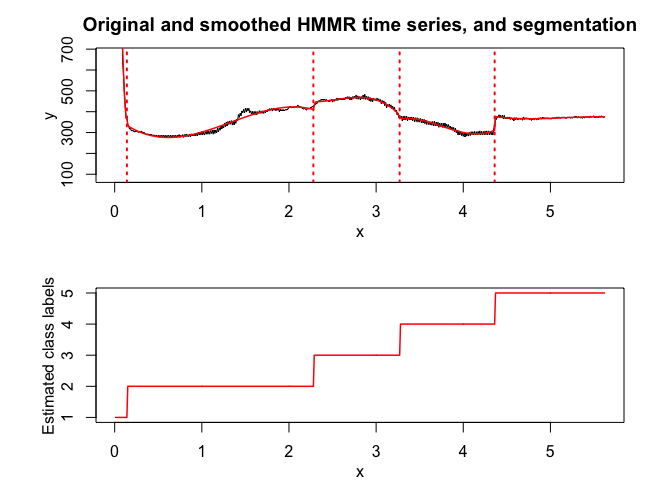
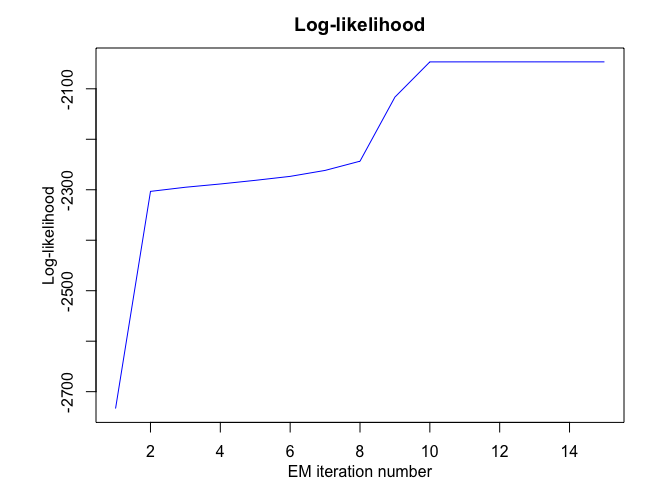
PWR
# Application to a toy data set
data("univtoydataset")
K <- 5 # Number of segments
p <- 1 # Polynomial degree
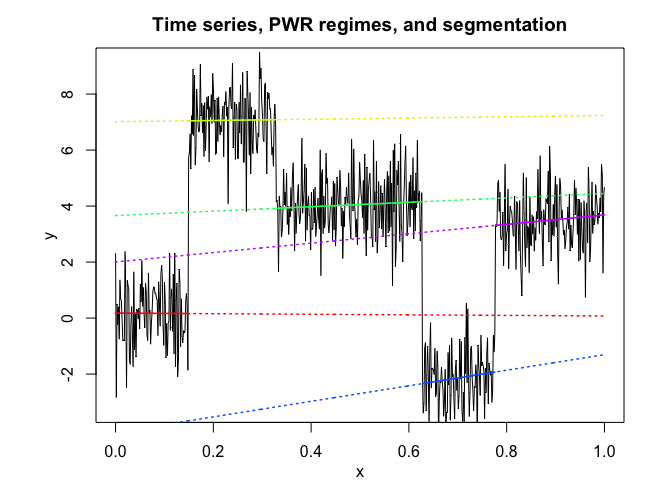
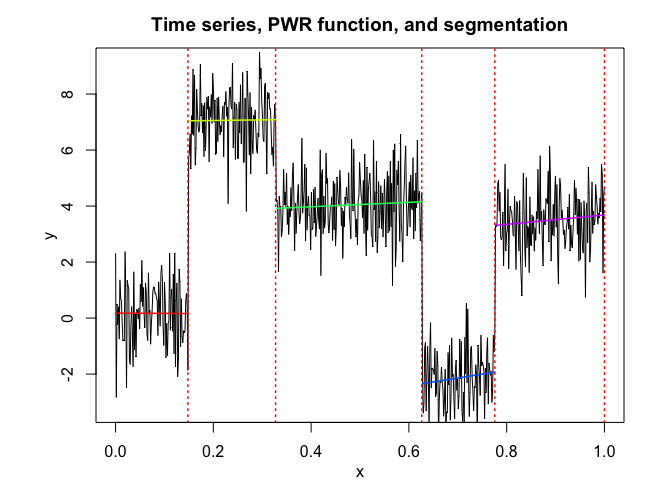
pwr <- fitPWRFisher(univtoydataset$x, univtoydataset$y, K, p)
pwr$plot()
# Application to a real data set
data("univrealdataset")
K <- 5 # Number of segments
p <- 3 # Polynomial degree
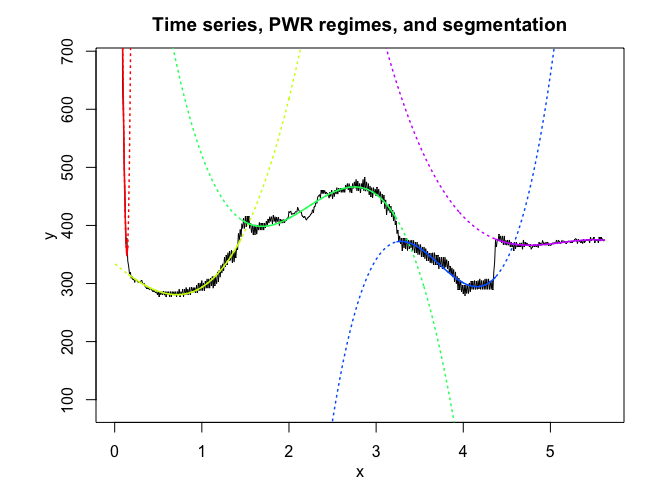
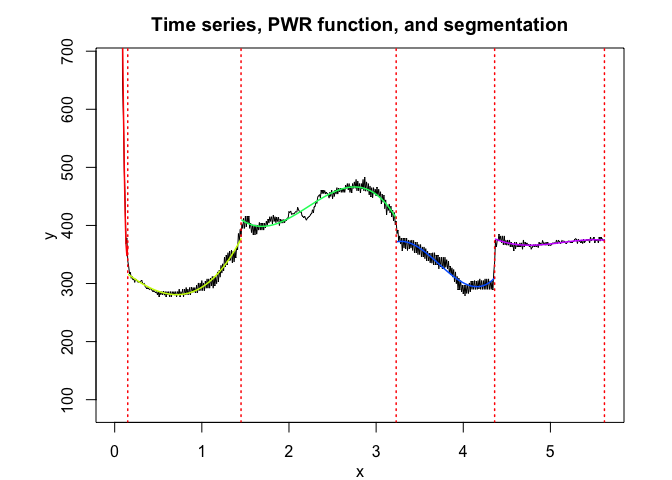
pwr <- fitPWRFisher(univrealdataset$x, univrealdataset$y2, K, p)
pwr$plot()
MRHLP
# Application to a toy data set
data("multivtoydataset")
K <- 5 # Number of regimes (mixture components)
p <- 1 # Dimension of beta (order of the polynomial regressors)
q <- 1 # Dimension of w (order of the logistic regression: to be set to 1 for segmentation)
variance_type <- "heteroskedastic" # "heteroskedastic" or "homoskedastic" model
n_tries <- 1
max_iter <- 1500
threshold <- 1e-6
verbose <- TRUE
verbose_IRLS <- FALSE
mrhlp <- emMRHLP(multivtoydataset$x, multivtoydataset[,c("y1", "y2", "y3")],
K, p, q, variance_type, n_tries, max_iter, threshold, verbose,
verbose_IRLS)
#> EM: Iteration : 1 || log-likelihood : -4807.6644322901
#> EM: Iteration : 2 || log-likelihood : -3314.25165556383
#> EM: Iteration : 3 || log-likelihood : -3216.8871750704
#> EM: Iteration : 4 || log-likelihood : -3126.33556053822
#> EM: Iteration : 5 || log-likelihood : -2959.59933830667
#> EM: Iteration : 6 || log-likelihood : -2895.65953485704
#> EM: Iteration : 7 || log-likelihood : -2892.93263500326
#> EM: Iteration : 8 || log-likelihood : -2889.34084959654
#> EM: Iteration : 9 || log-likelihood : -2884.56422084139
#> EM: Iteration : 10 || log-likelihood : -2878.29772085061
#> EM: Iteration : 11 || log-likelihood : -2870.61242183846
#> EM: Iteration : 12 || log-likelihood : -2862.86238149363
#> EM: Iteration : 13 || log-likelihood : -2856.85351443338
#> EM: Iteration : 14 || log-likelihood : -2851.74642203885
#> EM: Iteration : 15 || log-likelihood : -2850.00381259526
#> EM: Iteration : 16 || log-likelihood : -2849.86516522686
#> EM: Iteration : 17 || log-likelihood : -2849.7354103643
#> EM: Iteration : 18 || log-likelihood : -2849.56953544124
#> EM: Iteration : 19 || log-likelihood : -2849.40322468732
#> EM: Iteration : 20 || log-likelihood : -2849.40321381274
mrhlp$summary()
#> ----------------------
#> Fitted MRHLP model
#> ----------------------
#>
#> MRHLP model with K = 5 regimes
#>
#> log-likelihood nu AIC BIC ICL
#> -2849.403 68 -2917.403 -3070.651 -3069.896
#>
#> Clustering table:
#> 1 2 3 4 5
#> 100 120 200 100 150
#>
#>
#> ------------------
#> Regime 1 (K = 1):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 0.11943184 0.6087582 -2.038486
#> X^1 -0.08556857 4.1038126 2.540536
#>
#> Covariance matrix:
#>
#> 1.19063336 0.12765794 0.05537134
#> 0.12765794 0.87144062 -0.05213162
#> 0.05537134 -0.05213162 0.87885166
#> ------------------
#> Regime 2 (K = 2):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 6.924025 4.9368460 10.288339
#> X^1 1.118034 0.4726707 -1.409218
#>
#> Covariance matrix:
#>
#> 1.0690431 -0.18293369 0.12602459
#> -0.1829337 1.05280632 0.01390041
#> 0.1260246 0.01390041 0.75995058
#> ------------------
#> Regime 3 (K = 3):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 3.6535241 6.3654379 8.488318
#> X^1 0.6233579 -0.8866887 -1.126692
#>
#> Covariance matrix:
#>
#> 1.02591553 -0.05445227 -0.02019896
#> -0.05445227 1.18941700 0.01565240
#> -0.02019896 0.01565240 1.00257195
#> ------------------
#> Regime 4 (K = 4):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 -1.439637 -4.463014 2.952470
#> X^1 0.703211 3.649717 -4.187703
#>
#> Covariance matrix:
#>
#> 0.88000190 -0.03249118 -0.03411075
#> -0.03249118 1.12087583 -0.07881351
#> -0.03411075 -0.07881351 0.86060127
#> ------------------
#> Regime 5 (K = 5):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 3.4982408 2.5357751 7.652113
#> X^1 0.0574791 -0.7286824 -3.005802
#>
#> Covariance matrix:
#>
#> 1.13330209 0.25869951 0.03163467
#> 0.25869951 1.21230741 0.04746018
#> 0.03163467 0.04746018 0.80241715
mrhlp$plot()
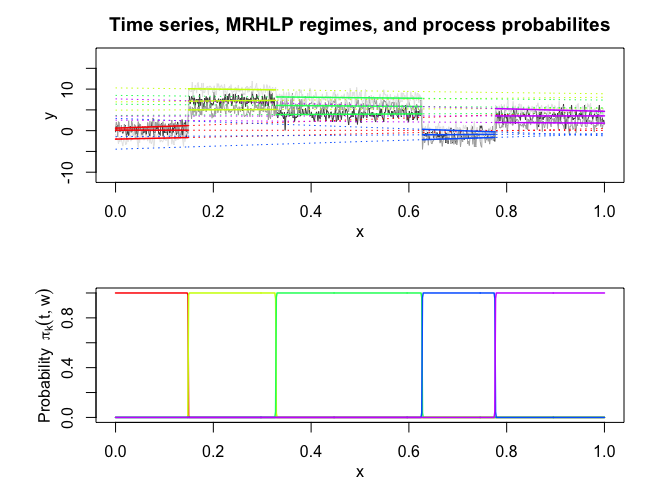
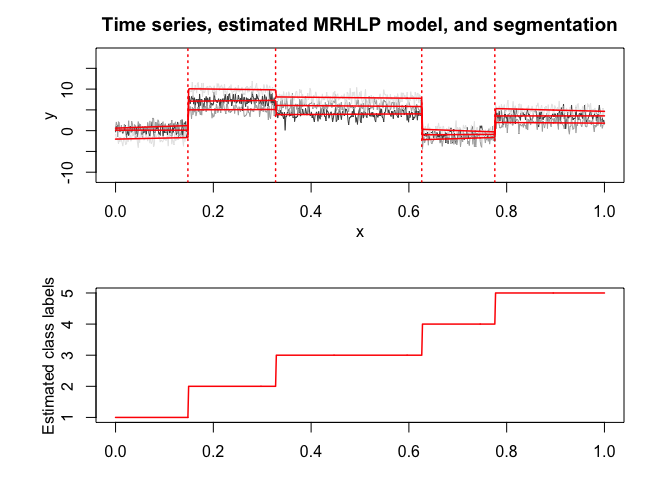
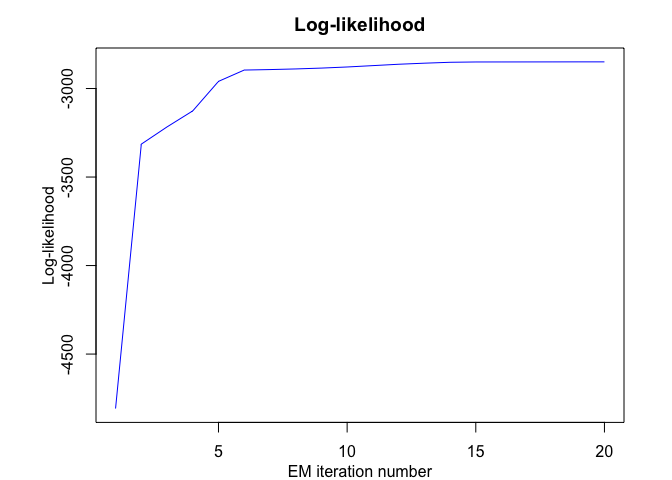
# Application to a real data set (human activity recogntion data)
data("multivrealdataset")
K <- 5 # Number of regimes (mixture components)
p <- 3 # Dimension of beta (order of the polynomial regressors)
q <- 1 # Dimension of w (order of the logistic regression: to be set to 1 for segmentation)
variance_type <- "heteroskedastic" # "heteroskedastic" or "homoskedastic" model
n_tries <- 1
max_iter <- 1500
threshold <- 1e-6
verbose <- TRUE
verbose_IRLS <- FALSE
mrhlp <- emMRHLP(multivrealdataset$x, multivrealdataset[,c("y1", "y2", "y3")],
K, p, q, variance_type, n_tries, max_iter, threshold, verbose,
verbose_IRLS)
#> EM: Iteration : 1 || log-likelihood : -792.888668727036
#> EM: Iteration : 2 || log-likelihood : 6016.45835957306
#> EM: Iteration : 3 || log-likelihood : 6362.81791662824
#> EM: Iteration : 4 || log-likelihood : 6615.72233403002
#> EM: Iteration : 5 || log-likelihood : 6768.32107943849
#> EM: Iteration : 6 || log-likelihood : 6840.97339565987
#> EM: Iteration : 7 || log-likelihood : 6860.97262839295
#> EM: Iteration : 8 || log-likelihood : 6912.25605673784
#> EM: Iteration : 9 || log-likelihood : 6945.96718258737
#> EM: Iteration : 10 || log-likelihood : 6951.28584396645
#> EM: Iteration : 11 || log-likelihood : 6952.37644678517
#> EM: Iteration : 12 || log-likelihood : 6954.80510338749
#> EM: Iteration : 13 || log-likelihood : 6958.99033092484
#> EM: Iteration : 14 || log-likelihood : 6964.81099837456
#> EM: Iteration : 15 || log-likelihood : 6999.90358068156
#> EM: Iteration : 16 || log-likelihood : 7065.39327246318
#> EM: Iteration : 17 || log-likelihood : 7166.23398344994
#> EM: Iteration : 18 || log-likelihood : 7442.73330846285
#> EM: Iteration : 19 || log-likelihood : 7522.65416438396
#> EM: Iteration : 20 || log-likelihood : 7524.41524338024
#> EM: Iteration : 21 || log-likelihood : 7524.57590110924
#> EM: Iteration : 22 || log-likelihood : 7524.73808801417
#> EM: Iteration : 23 || log-likelihood : 7524.88684996651
#> EM: Iteration : 24 || log-likelihood : 7524.9753964817
#> EM: Iteration : 25 || log-likelihood : 7524.97701548847
mrhlp$summary()
#> ----------------------
#> Fitted MRHLP model
#> ----------------------
#>
#> MRHLP model with K = 5 regimes
#>
#> log-likelihood nu AIC BIC ICL
#> 7524.977 98 7426.977 7146.696 7147.535
#>
#> Clustering table:
#> 1 2 3 4 5
#> 413 344 588 423 485
#>
#>
#> ------------------
#> Regime 1 (K = 1):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 1.64847721 2.33823068 9.40173242
#> X^1 -0.31396583 0.38235782 -0.10031616
#> X^2 0.23954454 -0.30105177 0.07812145
#> X^3 -0.04725267 0.06166899 -0.01586579
#>
#> Covariance matrix:
#>
#> 0.0200740364 -0.004238036 0.0004011388
#> -0.0042380363 0.006082904 -0.0012973026
#> 0.0004011388 -0.001297303 0.0013201963
#> ------------------
#> Regime 2 (K = 2):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 -106.0250571 -31.4671946 -107.9697464
#> X^1 45.2035210 21.2126134 72.0220177
#> X^2 -5.7330338 -4.1285514 -13.9857795
#> X^3 0.2343552 0.2485377 0.8374817
#>
#> Covariance matrix:
#>
#> 0.11899225 -0.03866052 -0.06693441
#> -0.03866052 0.17730401 0.04036629
#> -0.06693441 0.04036629 0.11983979
#> ------------------
#> Regime 3 (K = 3):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 9.0042249443 -1.247752962 -2.492119515
#> X^1 0.2191555621 0.418071041 0.310449523
#> X^2 -0.0242080660 -0.043802827 -0.039012607
#> X^3 0.0008494208 0.001474635 0.001427627
#>
#> Covariance matrix:
#>
#> 4.103351e-04 -0.0001330363 5.289199e-05
#> -1.330363e-04 0.0006297205 2.027763e-04
#> 5.289199e-05 0.0002027763 1.374405e-03
#> ------------------
#> Regime 4 (K = 4):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 -1029.9071752 334.4975068 466.0981076
#> X^1 199.9531885 -68.7252041 -105.6436899
#> X^2 -12.6550086 4.6489685 7.6555642
#> X^3 0.2626998 -0.1032161 -0.1777453
#>
#> Covariance matrix:
#>
#> 0.058674116 -0.017661572 0.002139975
#> -0.017661572 0.047588713 0.007867532
#> 0.002139975 0.007867532 0.067150809
#> ------------------
#> Regime 5 (K = 5):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 27.247199195 -14.393798357 19.741283724
#> X^1 -3.530625667 2.282492947 -1.511225702
#> X^2 0.161234880 -0.101613670 0.073003292
#> X^3 -0.002446104 0.001490288 -0.001171127
#>
#> Covariance matrix:
#>
#> 6.900384e-03 -0.001176838 2.966199e-05
#> -1.176838e-03 0.003596238 -2.395420e-04
#> 2.966199e-05 -0.000239542 5.573451e-04
mrhlp$plot()
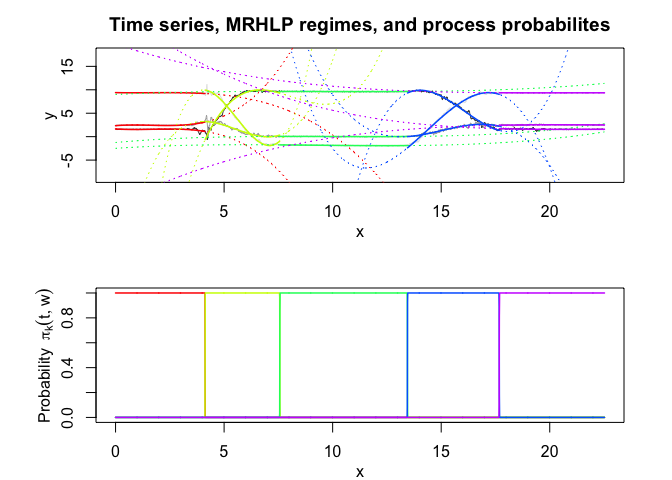
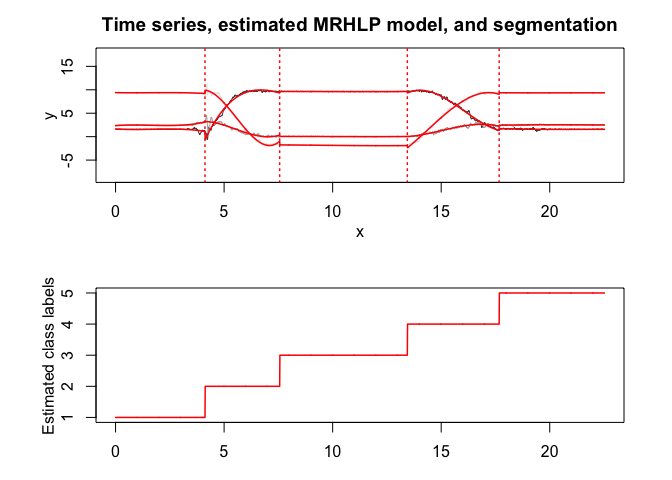
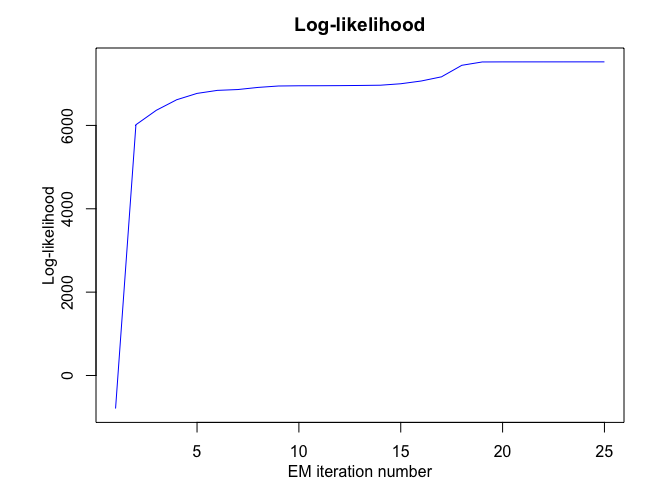
MHMMR
# Application to a simulated data set
data("multivtoydataset")
K <- 5 # Number of regimes (states)
p <- 1 # Dimension of beta (order of the polynomial regressors)
variance_type <- "heteroskedastic" # "heteroskedastic" or "homoskedastic" model
n_tries <- 1
max_iter <- 1500
threshold <- 1e-6
verbose <- TRUE
mhmmr <- emMHMMR(multivtoydataset$x, multivtoydataset[, c("y1", "y2", "y3")],
K, p, variance_type, n_tries, max_iter, threshold, verbose)
#> EM: Iteration : 1 || log-likelihood : -4539.37845473736
#> EM: Iteration : 2 || log-likelihood : -3075.7862970485
#> EM: Iteration : 3 || log-likelihood : -2904.71126233611
#> EM: Iteration : 4 || log-likelihood : -2883.23456594806
#> EM: Iteration : 5 || log-likelihood : -2883.12446634454
#> EM: Iteration : 6 || log-likelihood : -2883.12436399888
mhmmr$summary()
#> ----------------------
#> Fitted MHMMR model
#> ----------------------
#>
#> MHMMR model with K = 5 regimes
#>
#> log-likelihood nu AIC BIC
#> -2883.124 84 -2967.124 -3156.43
#>
#> Clustering table:
#> 1 2 3 4 5
#> 100 120 200 100 150
#>
#>
#> ------------------
#> Regime 1 (K = 1):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 0.11943184 0.6087582 -2.038486
#> X^1 -0.08556857 4.1038126 2.540536
#>
#> Covariance matrix:
#>
#> 1.19064336 0.12765794 0.05537134
#> 0.12765794 0.87145062 -0.05213162
#> 0.05537134 -0.05213162 0.87886166
#> ------------------
#> Regime 2 (K = 2):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 6.921139 4.9377164 10.290536
#> X^1 1.131946 0.4684922 -1.419758
#>
#> Covariance matrix:
#>
#> 1.0688949 -0.18240787 0.12675972
#> -0.1824079 1.05317924 0.01419686
#> 0.1267597 0.01419686 0.76030310
#> ------------------
#> Regime 3 (K = 3):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 3.6576562 6.3642526 8.493765
#> X^1 0.6155173 -0.8844373 -1.137027
#>
#> Covariance matrix:
#>
#> 1.02647251 -0.05491451 -0.01930098
#> -0.05491451 1.18921808 0.01510035
#> -0.01930098 0.01510035 1.00352482
#> ------------------
#> Regime 4 (K = 4):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 -1.439637 -4.463014 2.952470
#> X^1 0.703211 3.649717 -4.187703
#>
#> Covariance matrix:
#>
#> 0.88001190 -0.03249118 -0.03411075
#> -0.03249118 1.12088583 -0.07881351
#> -0.03411075 -0.07881351 0.86061127
#> ------------------
#> Regime 5 (K = 5):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 3.4982408 2.5357751 7.652113
#> X^1 0.0574791 -0.7286824 -3.005802
#>
#> Covariance matrix:
#>
#> 1.13331209 0.25869951 0.03163467
#> 0.25869951 1.21231741 0.04746018
#> 0.03163467 0.04746018 0.80242715
mhmmr$plot(what = c("smoothed", "regressors", "loglikelihood"))
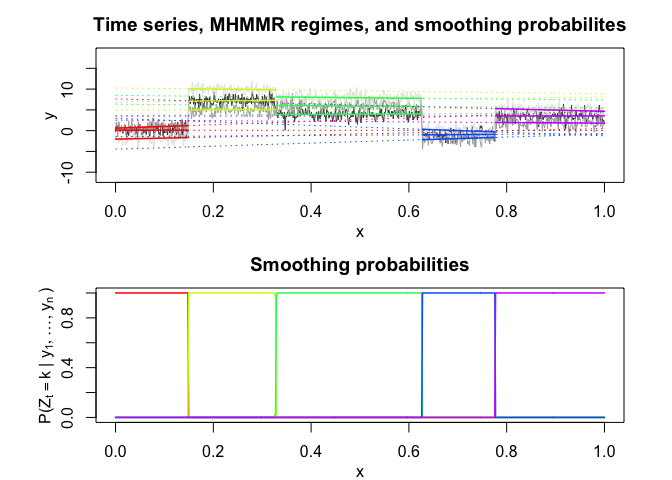
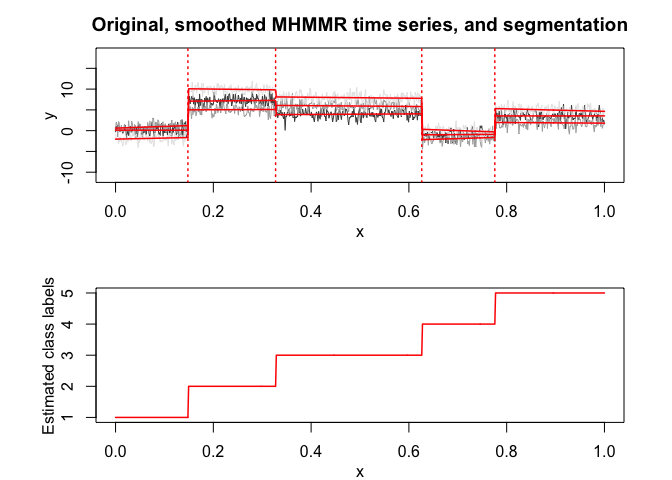
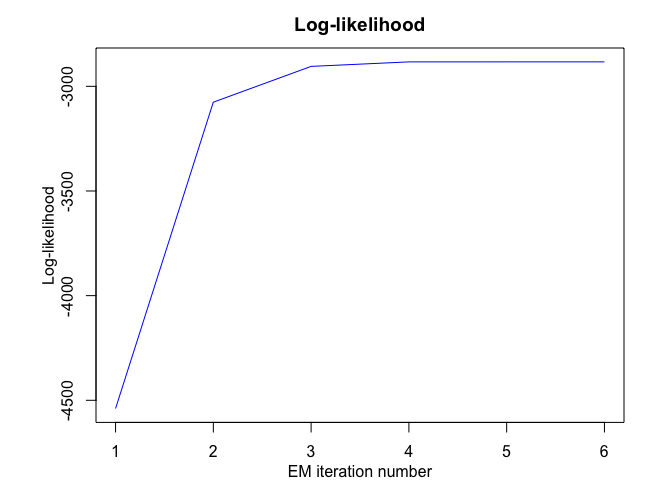
# Application to a real data set (human activity recognition data)
data("multivrealdataset")
K <- 5 # Number of regimes (states)
p <- 3 # Dimension of beta (order of the polynomial regressors)
variance_type <- "heteroskedastic" # "heteroskedastic" or "homoskedastic" model
n_tries <- 1
max_iter <- 1500
threshold <- 1e-6
verbose <- TRUE
mhmmr <- emMHMMR(multivrealdataset$x, multivrealdataset[, c("y1", "y2", "y3")],
K, p, variance_type, n_tries, max_iter, threshold, verbose)
#> EM: Iteration : 1 || log-likelihood : 817.206309249687
#> EM: Iteration : 2 || log-likelihood : 1793.49320726452
#> EM: Iteration : 3 || log-likelihood : 1908.47251424374
#> EM: Iteration : 4 || log-likelihood : 2006.7976746047
#> EM: Iteration : 5 || log-likelihood : 3724.91911814713
#> EM: Iteration : 6 || log-likelihood : 3846.02584774854
#> EM: Iteration : 7 || log-likelihood : 3957.04953794437
#> EM: Iteration : 8 || log-likelihood : 4008.60804596975
#> EM: Iteration : 9 || log-likelihood : 4011.09964067314
#> EM: Iteration : 10 || log-likelihood : 4014.35810165377
#> EM: Iteration : 11 || log-likelihood : 4026.38632031497
#> EM: Iteration : 12 || log-likelihood : 4027.13758668835
#> EM: Iteration : 13 || log-likelihood : 4027.13639613206
mhmmr$summary()
#> ----------------------
#> Fitted MHMMR model
#> ----------------------
#>
#> MHMMR model with K = 5 regimes
#>
#> log-likelihood nu AIC BIC
#> 4027.136 114 3913.136 3587.095
#>
#> Clustering table:
#> 1 2 3 4 5
#> 461 297 587 423 485
#>
#>
#> ------------------
#> Regime 1 (K = 1):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 1.41265303 2.42222746 9.381994682
#> X^1 0.47242692 0.09217574 -0.023282898
#> X^2 -0.28135064 -0.10169173 0.018998710
#> X^3 0.04197568 0.02620151 -0.004217078
#>
#> Covariance matrix:
#>
#> 0.12667921 -0.019381009 -0.018810846
#> -0.01938101 0.109202105 -0.001402791
#> -0.01881085 -0.001402791 0.026461790
#> ------------------
#> Regime 2 (K = 2):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 -3.6868321 2.4724043 7.794639
#> X^1 -6.8471097 4.6786664 14.749215
#> X^2 2.9742521 -1.4716819 -4.646020
#> X^3 -0.2449644 0.1076065 0.335142
#>
#> Covariance matrix:
#>
#> 0.22604244 -0.032716477 0.013626769
#> -0.03271648 0.032475350 0.008585402
#> 0.01362677 0.008585402 0.041960228
#> ------------------
#> Regime 3 (K = 3):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 0.776245522 0.014437427 -0.1144683124
#> X^1 2.627158141 0.048519275 -0.3883099866
#> X^2 -0.255314738 -0.008318957 0.0283047828
#> X^3 0.008129981 0.000356239 -0.0007003718
#>
#> Covariance matrix:
#>
#> 0.0012000978 -0.0002523608 -0.0001992900
#> -0.0002523608 0.0006584694 0.0002391577
#> -0.0001992900 0.0002391577 0.0014228769
#> ------------------
#> Regime 4 (K = 4):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 0.002894474 -0.0002900823 -0.001513232
#> X^1 0.029936273 -0.0029993910 -0.015647636
#> X^2 0.232798943 -0.0233058753 -0.121611904
#> X^3 -0.013209774 0.0019141508 0.009151938
#>
#> Covariance matrix:
#>
#> 0.21455830 -0.07328139 -0.08824736
#> -0.07328139 0.17055704 0.45218611
#> -0.08824736 0.45218611 1.76616982
#> ------------------
#> Regime 5 (K = 5):
#>
#> Regression coefficients:
#>
#> Beta(d = 1) Beta(d = 2) Beta(d = 3)
#> 1 9.416685e-05 0.0001347198 0.0005119141
#> X^1 1.259159e-03 0.0018014389 0.0068451694
#> X^2 1.265758e-02 0.0181095390 0.0688126905
#> X^3 -4.344666e-04 -0.0005920827 -0.0022723501
#>
#> Covariance matrix:
#>
#> 0.009259719 -0.000696446 0.006008102
#> -0.000696446 0.003732296 0.001056145
#> 0.006008102 0.001056145 0.016144263
mhmmr$plot(what = c("smoothed", "regressors", "loglikelihood"))
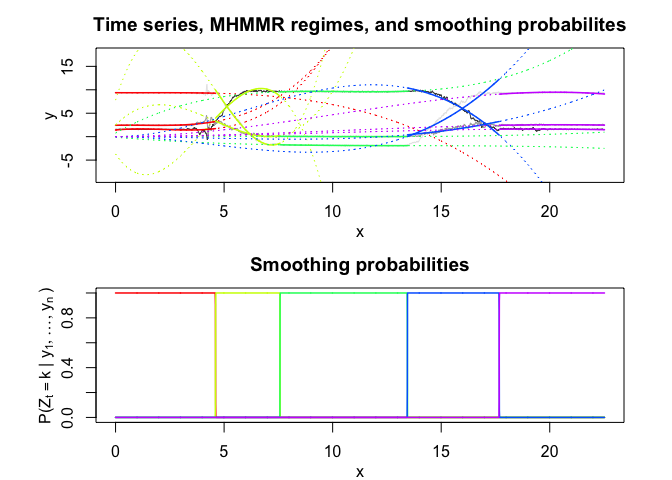
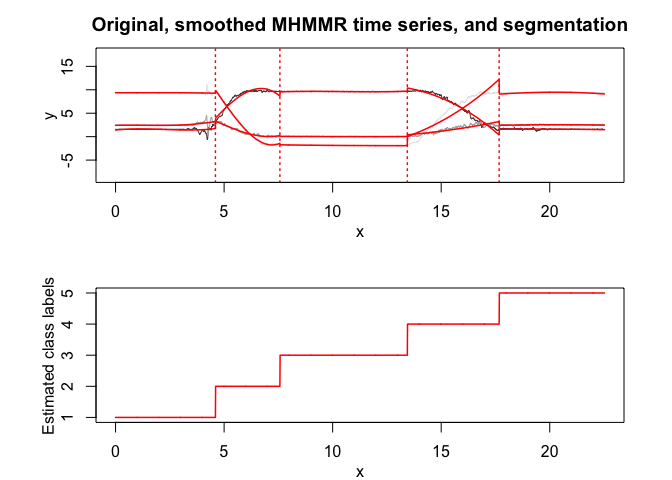
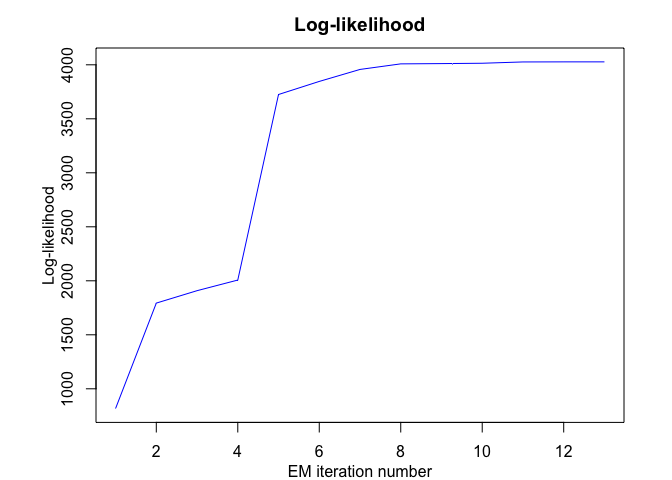
Model selection
In this package, it is possible to select models based on information criteria such as BIC, AIC and ICL.
The selection can be done for the two following parameters:
- K: The number of regimes;
- p: The order of the polynomial regression.
RHLP
Let’s select a RHLP model for the following time series Y:
data("univtoydataset")
x = univtoydataset$x
y = univtoydataset$y
plot(x, y, type = "l", xlab = "x", ylab = "Y")
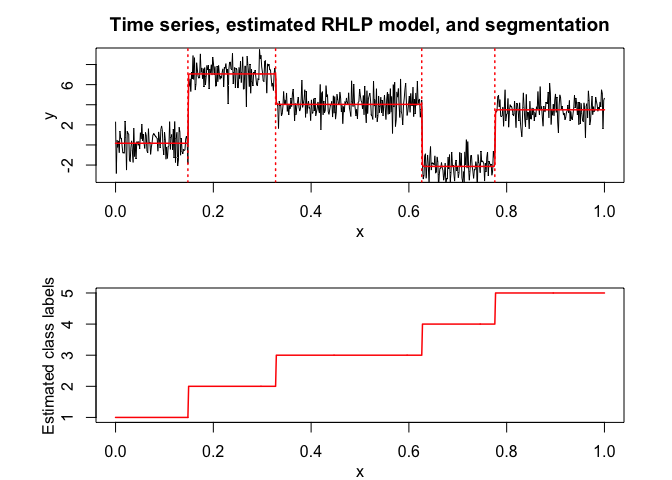
selectedrhlp <- selectRHLP(X = x, Y = y, Kmin = 2, Kmax = 6, pmin = 0, pmax = 3)
#> The RHLP model selected via the "BIC" has K = 5 regimes
#> and the order of the polynomial regression is p = 0.
#> BIC = -1041.40789532438
#> AIC = -1000.84239591291
selectedrhlp$plot(what = "estimatedsignal")
HMMR
Let’s select a HMMR model for the following time series Y:
data("univtoydataset")
x = univtoydataset$x
y = univtoydataset$y
plot(x, y, type = "l", xlab = "x", ylab = "Y")
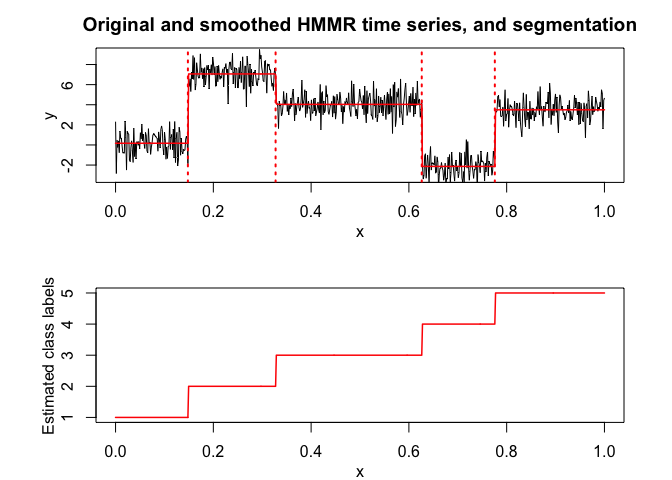
selectedhmmr <- selectHMMR(X = x, Y = y, Kmin = 2, Kmax = 6, pmin = 0, pmax = 3)
#> The HMMR model selected via the "BIC" has K = 5 regimes
#> and the order of the polynomial regression is p = 0.
#> BIC = -1136.39152222095
#> AIC = -1059.76780111041
selectedhmmr$plot(what = "smoothed")
MRHLP
Let’s select a MRHLP model for the following multivariate time series Y:
data("multivtoydataset")
x <- multivtoydataset$x
y <- multivtoydataset[, c("y1", "y2", "y3")]
matplot(x, y, type = "l", xlab = "x", ylab = "Y", lty = 1)
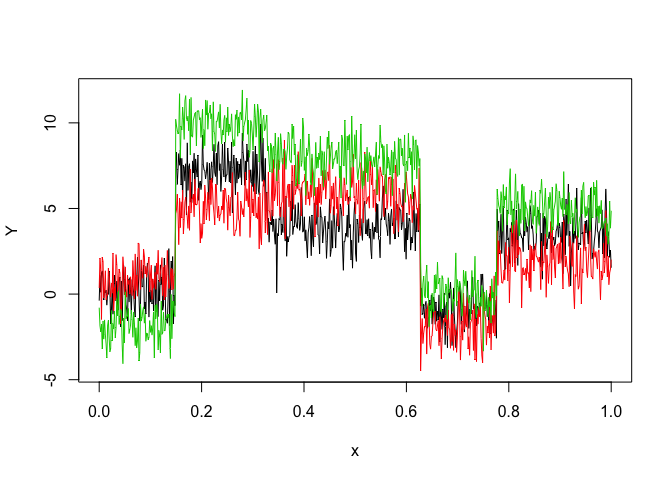
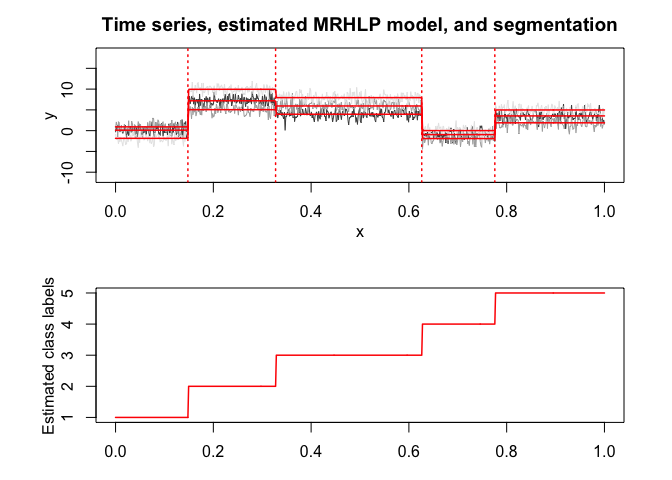
selectedmrhlp <- selectMRHLP(X = x, Y = y, Kmin = 2, Kmax = 6, pmin = 0, pmax = 3)
#> Warning in emMRHLP(X = X1, Y = Y1, K, p): EM log-likelihood is decreasing
#> from -3105.78591044952to -3105.78627830471 !
#> The MRHLP model selected via the "BIC" has K = 5 regimes
#> and the order of the polynomial regression is p = 0.
#> BIC = -3033.20042397111
#> AIC = -2913.75756459291
selectedmrhlp$plot(what = "estimatedsignal")
MHMMR
Let’s select a MHMMR model for the following multivariate time series Y:
data("multivtoydataset")
x <- multivtoydataset$x
y <- multivtoydataset[, c("y1", "y2", "y3")]
matplot(x, y, type = "l", xlab = "x", ylab = "Y", lty = 1)
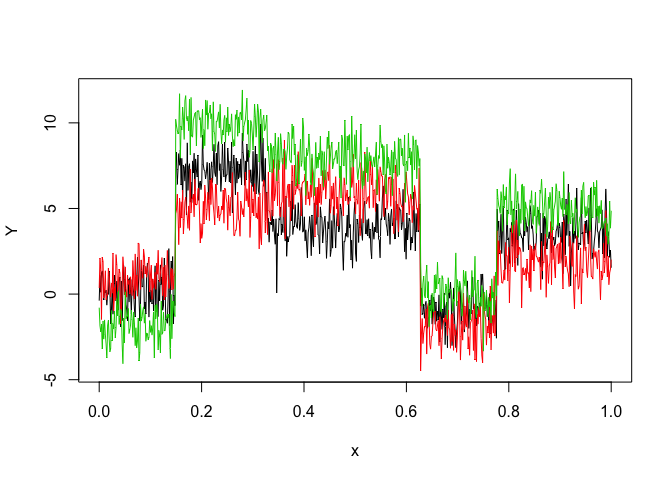
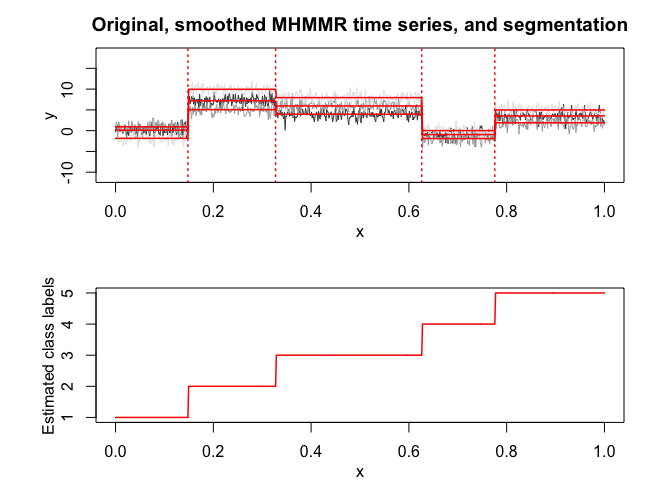
selectedmhmmr <- selectMHMMR(X = x, Y = y, Kmin = 2, Kmax = 6, pmin = 0, pmax = 3)
#> The MHMMR model selected via the "BIC" has K = 5 regimes
#> and the order of the polynomial regression is p = 0.
#> BIC = -3118.9815385353
#> AIC = -2963.48045745801
selectedmhmmr$plot(what = "smoothed")
References
Chamroukhi, Faicel, and Hien D. Nguyen. 2019. “Model-Based Clustering and Classification of Functional Data.” Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery. https://chamroukhi.com/papers/MBCC-FDA.pdf.
Chamroukhi, F. 2015. “Statistical Learning of Latent Data Models for Complex Data Analysis.” Habilitation Thesis (HDR), Université de Toulon. https://chamroukhi.com/Dossier/FChamroukhi-Habilitation.pdf.
Trabelsi, D., S. Mohammed, F. Chamroukhi, L. Oukhellou, and Y. Amirat. 2013. “An Unsupervised Approach for Automatic Activity Recognition Based on Hidden Markov Model Regression.” IEEE Transactions on Automation Science and Engineering 3 (10): 829–335. https://chamroukhi.com/papers/Chamroukhi-MHMMR-IeeeTase.pdf.
Chamroukhi, F., D. Trabelsi, S. Mohammed, L. Oukhellou, and Y. Amirat. 2013. “Joint Segmentation of Multivariate Time Series with Hidden Process Regression for Human Activity Recognition.” Neurocomputing 120: 633–44. https://chamroukhi.com/papers/chamroukhi_et_al_neucomp2013b.pdf.
Chamroukhi, F., A. Samé, G. Govaert, and P. Aknin. 2010. “A Hidden Process Regression Model for Functional Data Description. Application to Curve Discrimination.” Neurocomputing 73 (7-9): 1210–21. https://chamroukhi.com/papers/chamroukhi_neucomp_2010.pdf.
Chamroukhi, F. 2010. “Hidden Process Regression for Curve Modeling, Classification and Tracking.” Ph.D. Thesis, Université de Technologie de Compiègne. https://chamroukhi.com/papers/FChamroukhi-Thesis.pdf.
Chamroukhi, F., A. Samé, G. Govaert, and P. Aknin. 2009. “Time Series Modeling by a Regression Approach Based on a Latent Process.” Neural Networks 22 (5-6): 593–602. https://chamroukhi.com/papers/Chamroukhi_Neural_Networks_2009.pdf.