Fitting Shared Atoms Nested Models via MCMC or Variational Bayes.
sanba v0.0.1
The goal of sanba is to estimate Bayesian nested mixture models via MCMC and VI methods. Specifically, the package implements the common atoms model (Denti et al., 2023) and hybrid finite-infinite models (D’Angelo and Denti, 2024). All models use Gaussian mixtures with a normal-inverse-gamma prior distribution on the parameters. Additional functions are provided to help analyzing the results of the fitting procedure.
Installation
You can install the development version of sanba from GitHub with:
# install.packages("devtools")
devtools::install_github("Fradenti/sanba")
Examples
Fitting via MCMC
library(sanba)
#> Loading required package: scales
#> Loading required package: RColorBrewer
set.seed(123)
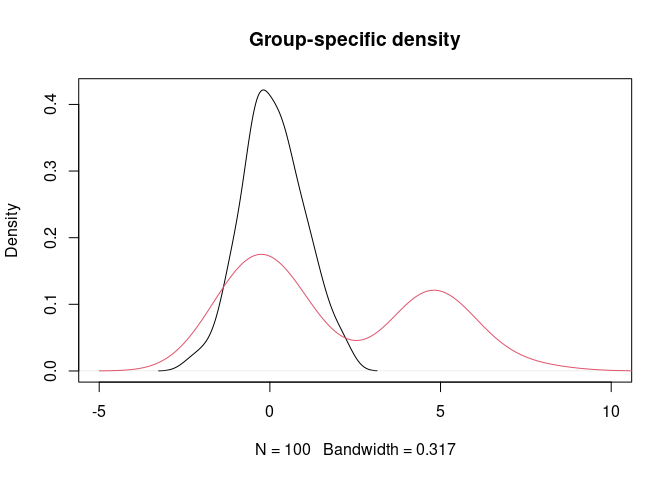
y <- c(rnorm(160), rnorm(40, 5))
g <- rep(1:2, rep(100, 2))
plot(density(y[g==1]), xlim = c(-5,10), main = "Group-specific density")
lines(density(y[g==2]), col = 2)
out_mcmc <- fit_CAM(y = y, group = g, est_method = "MCMC", mcmc_param = list(nrep = 5000, burn=3000))
#> Warning in sample_CAM(y, group, prior_param = prior_param, mcmc_param =
#> mcmc_param): Increase maxK: all the provided distributional mixture components
#> were used. Check '$warnings' to see when it happened.
out_mcmc
#>
#> MCMC result of CAM model
#> -----------------------------------------------
#> Model estimated on 200 total observations and 2 groups
#> Size of the MCMC sample (after burn-in): 2000
#> Total MCMC iterations performed: 5000
#> maxL: 50 - maxK: 20
#> Elapsed time: 0.615 secs
plot(out_mcmc)
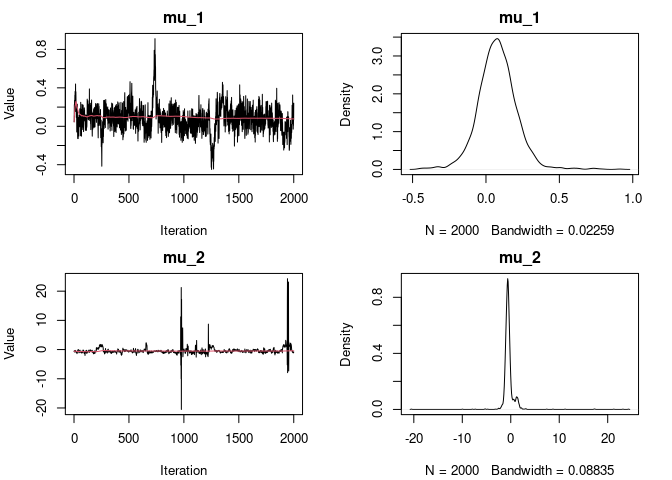
#> [1] "Output truncated at 2 for mu."
Fitting via VI
library(sanba)
set.seed(123)
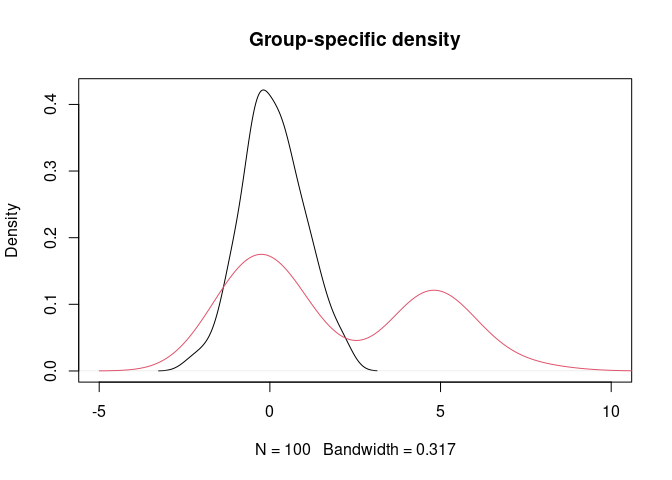
y <- c(rnorm(160), rnorm(40, 5))
g <- rep(1:2, rep(100, 2))
plot(density(y[g==1]), xlim = c(-5,10), main = "Group-specific density")
lines(density(y[g==2]), col = 2)
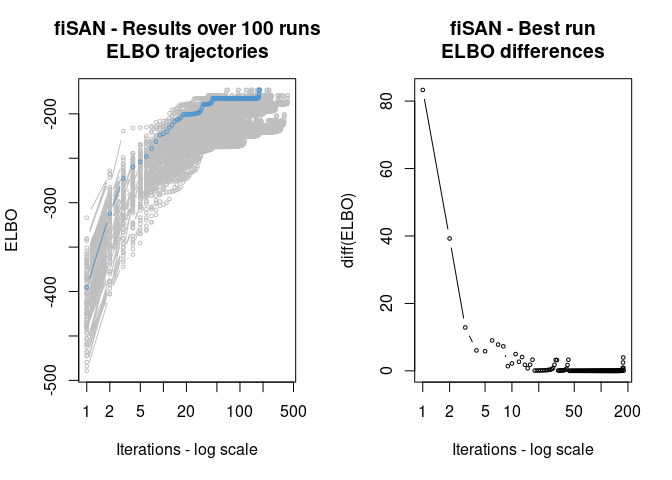
out_vi <- fit_fiSAN(y, group = g, est_method = "VI", vi_param = list(n_runs = 100))
out_vi
#> Variational inference results for fiSAN
#> ----------------------------------------------
#> L: 30 - K: 20
#> Threshold: 1e-06
#> ELBO value: -173.141
#> Best run out of 100
#> Convergence reached in 180 iterations
#> Elapsed time: 0.041 secs
plot(out_vi)
References
D’Angelo, L., and Denti, F. (2024). A Finite-Infinite Shared Atoms Nested Model for the Bayesian Analysis of Large Grouped Data Sets. Bayesian Analysis
Denti, F., Camerlenghi, F., Guindani, M., Mira, A., 2023. A Common Atoms Model for the Bayesian Nonparametric Analysis of Nested Data. Journal of the American Statistical Association. 118(541), 405–416.
