Description
Access and Format Single-Cell RNA-Seq Datasets from Public Resources.
Description
The goal of 'scfetch' is to access and format single-cell RNA-seq datasets. It can be used to download single-cell RNA-seq datasets from widely used public resources, including GEO <https://www.ncbi.nlm.nih.gov/geo/>, Zenodo <https://zenodo.org/>, CELLxGENE <https://cellxgene.cziscience.com/>, Human Cell Atlas <https://www.humancellatlas.org/>, PanglaoDB <https://panglaodb.se/index.html> and UCSC Cell Browser <https://cells.ucsc.edu/>. And, it can also be used to perform object conversion between SeuratObject <https://satijalab.org/seurat/>, loom <http://loompy.org/>, h5ad <https://scanpy.readthedocs.io/en/stable/>, SingleCellExperiment <https://bioconductor.org/packages/release/bioc/html/scran.html>, CellDataSet <http://cole-trapnell-lab.github.io/monocle-release/> and cell_data_set <https://cole-trapnell-lab.github.io/monocle3/>.
README.md
scfetch - Access and Format Single-cell RNA-seq Datasets from Public Resources
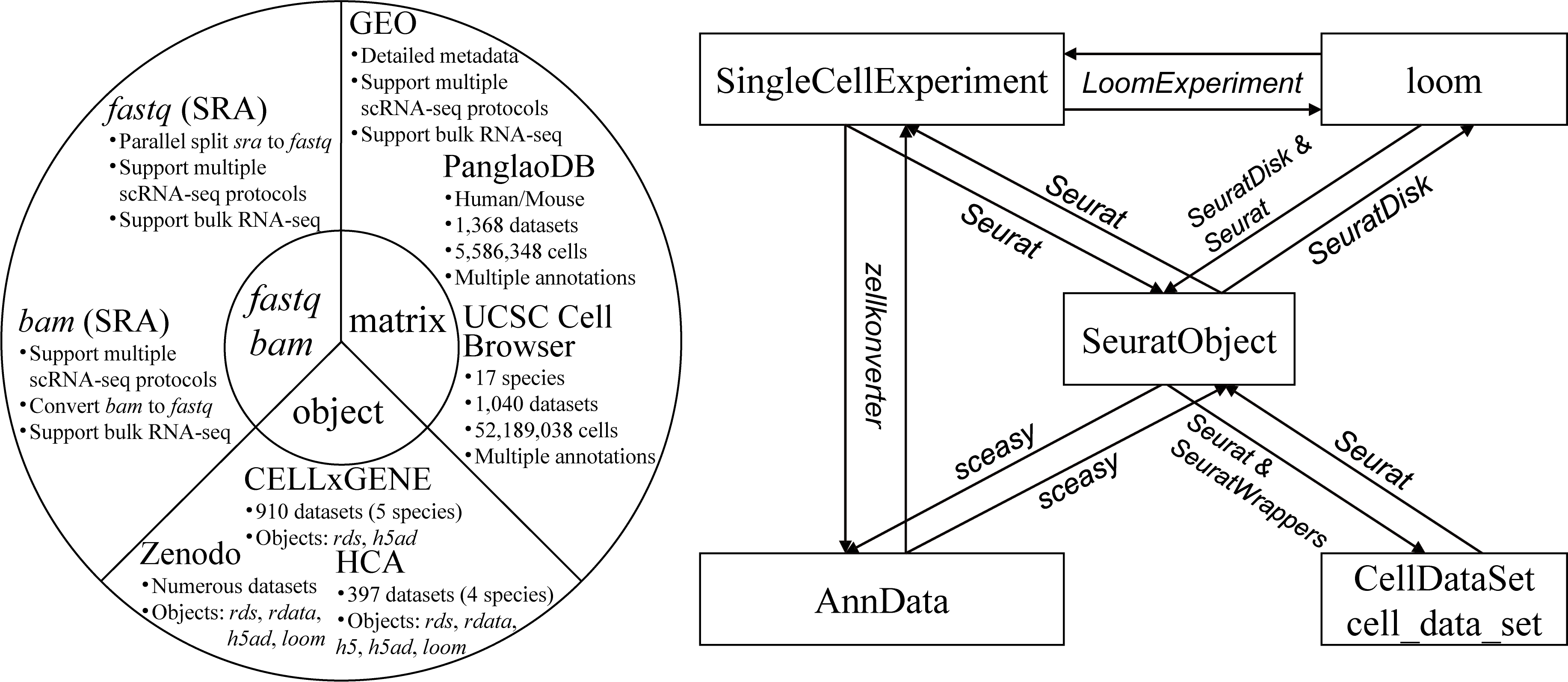
Introduction
scfetch is designed to accelerate users download and prepare single-cell datasets from public resources. It can be used to:
- Download fastq files from
GEO/SRA, foramt fastq files to standard style that can be identified by 10x softwares (e.g. CellRanger). - Download bam files from
GEO/SRA, support downloading original 10x generated bam files (with custom tags) and normal bam files, and convert bam files to fastq files. - Download scRNA-seq matrix and annotation (e.g. cell type) information from
GEO,PanglanDBandUCSC Cell Browser, load the downnloaded matrix toSeurat. - Download processed objects from
Zeenodo,CELLxGENEandHuman Cell Atlas. - Formats conversion between widely used single cell objects (
SeuratObject,AnnData,SingleCellExperiment,CellDataSet/cell_data_setandloom).
Framework
Installation
You can install the development version of scfetch from GitHub with:
# install.packages("devtools")
devtools::install_github("showteeth/scfetch")
For issues about installation, please refer INSTALL.md.
For data structures conversion, scfetch requires several python pcakages, you can install with:
# install python packages
conda install -c bioconda loompy anndata
# or
pip install anndata loompy
Usage
Vignette
Detailed usage is available in website.
Function list
| Type | Function | Usage |
|---|---|---|
| Download and format fastq | ExtractRun | Extract runs with GEO accession number or GSM number |
| DownloadSRA | Download sra files | |
| SplitSRA | Split sra files to fastq files and format to 10x standard style | |
| Download and convert bam | DownloadBam | Download bam (support 10x original bam) |
| Bam2Fastq | Convert bam files to fastq files | |
| Download matrix and load to Seurat | ExtractGEOMeta | Extract sample metadata from GEO |
| ParseGEO | Download matrix from GEO and load to Seurat | |
| ExtractPanglaoDBMeta | Extract sample metadata from PandlaoDB | |
| ExtractPanglaoDBComposition | Extract cell type composition of PanglaoDB datasets | |
| ParsePanglaoDB | Download matrix from PandlaoDB and load to Seurat | |
| ShowCBDatasets | Show all available datasets in UCSC Cell Browser | |
| ExtractCBDatasets | Extract UCSC Cell Browser datasets with attributes | |
| ExtractCBComposition | Extract cell type composition of UCSC Cell Browser datasets | |
| ParseCBDatasets | Download UCSC Cell Browser datasets and load to Seurat | |
| Download objects | ExtractZenodoMeta | Extract sample metadata from Zenodo with DOIs |
| ParseZenodo | Download rds/rdata/h5ad/loom from Zenodo with DOIs | |
| ShowCELLxGENEDatasets | Show all available datasets in CELLxGENE | |
| ExtractCELLxGENEMeta | Extract metadata of CELLxGENE datasets with attributes | |
| ParseCELLxGENE | Download rds/h5ad from CELLxGENE | |
| ShowHCAProjects | Show all available projects in Human Cell Atlas | |
| ExtractHCAMeta | Extract metadata of Human Cell Atlas projects with attributes | |
| ParseHCA | Download rds/rdata/h5/h5ad/loom from Human Cell Atlas | |
| Convert between different single-cell objects | ExportSeurat | Convert SeuratObject to AnnData, SingleCellExperiment, CellDataSet/cell_data_set and loom |
| ImportSeurat | Convert AnnData, SingleCellExperiment, CellDataSet/cell_data_set and loom to SeuratObject | |
| SCEAnnData | Convert between SingleCellExperiment and AnnData | |
| SCELoom | Convert between SingleCellExperiment and loom | |
| Summarize datasets based on attributes | StatDBAttribute | Summarize datasets in PandlaoDB, UCSC Cell Browser and CELLxGENE based on attributes |
Contact
For any question, feature request or bug report please write an email to [email protected].
Code of Conduct
Please note that the scfetch project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.