Description
A Single Cell Transcriptomics Based Deconvolution Pipeline for Leukemia.
Description
Given a bulk transcriptomic (RNA-seq) sample of an Myeloid Leukemia patient calculates immune composition and drug resistance for different small-molecule inhibitors. Published in <https://www.nature.com/articles/s41698-024-00596-9>.
README.md
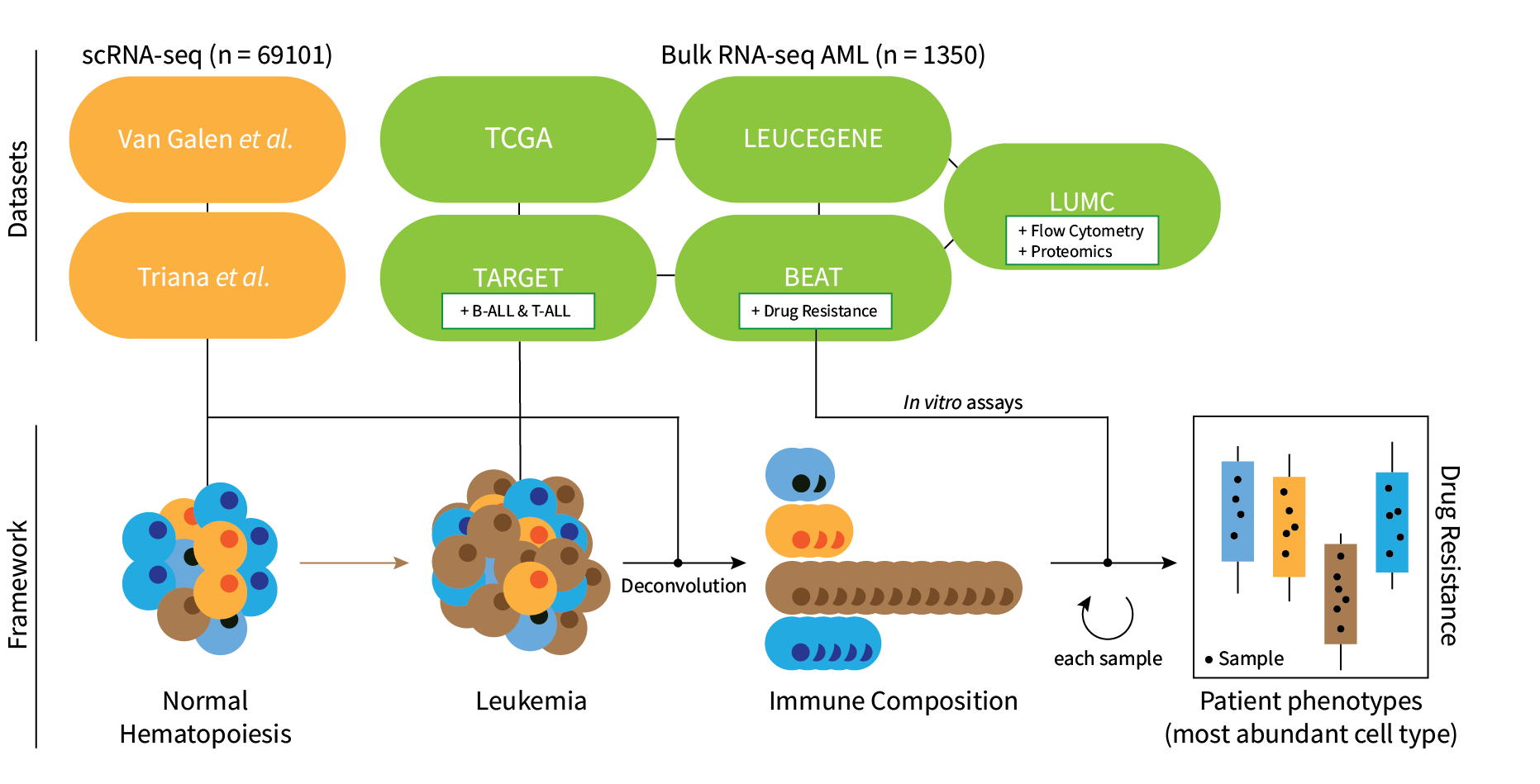
seAMLess
Overview
seAMLess is a wrapper function which deconvolutes bulk Acute Myeloid Leukemia (AML) RNA-seq samples with a healthy single cell reference atlas.
Installation
# CRAN mirror
install.packages("seAMLess")
Development version
To get bug fix and use a feature from the development version:
# install.packages("devtools")
devtools::install_github("eonurk/seAMLess")
Bioconda
seAMLess is also available in Bioconda and can be installed via:
conda install -c bioconda r-seamless
Next, install the seAMLessData package inside of R, this will take a few minutes:
install.packages("seAMLessData", repos = "https://eonurk.github.io/drat/")
Usage
library(seAMLess)
library(xbioc) # required
data(exampleTCGA)
head(exampleTCGA)[,1:4]
## X TCGA.AB.2856.03A TCGA.AB.2849.03A TCGA.AB.2971.03A
## 1 ENSG00000000003.13 7 9 1
## 2 ENSG00000000005.5 0 1 0
## 3 ENSG00000000419.11 689 661 555
## 4 ENSG00000000457.12 633 1434 855
## 5 ENSG00000000460.15 372 1211 519
## 6 ENSG00000000938.11 14712 405 6076
# Now run the function
res <- seAMLess(exampleTCGA)
## >> Human ensembl ids are converted to symbols...
## >> Deconvoluting samples...
## >> Deconvolution completed...
## >> Predicting Venetoclax resistance...
## >> Done...
# AML deconvolution
head(res$Deconvolution)[,1:4]
## CD14 Mono GMP T Cells pre B
## TCGA.AB.2856.03A 0.1495886 0.7079107 0.022868623 0.00000000
## TCGA.AB.2849.03A 0.0000000 0.0000000 0.000000000 0.00000000
## TCGA.AB.2971.03A 0.5494418 0.4462562 0.002898678 0.00000000
## TCGA.AB.2930.03A 0.0000000 0.4698555 0.000000000 0.00000000
## TCGA.AB.2891.03A 0.0000000 0.6189622 0.018645484 0.01384012
## TCGA.AB.2872.03A 0.0000000 0.9950150 0.000000000 0.00000000
# Create ternary plot
ternaryPlot(res)
# Venetoclax resistance
res$Venetoclax.resistance[1:4]
## TCGA.AB.2856.03A TCGA.AB.2849.03A TCGA.AB.2971.03A TCGA.AB.2930.03A
## 0.5070113 0.3242576 0.6678995 0.3305996
Contribution
You can send pull requests to make your contributions.
Author
License
- GNU General Public License v3.0