Handling of Data from the Clinical Data Management System 'secuTrial'.
secuTrialR
An R package to handle data from the clinical data management system (CDMS) secuTrial.
Installing from GitHub with devtools
Please note that R versions >= 3.5 should be used to run secuTrialR.
devtools::install_github("SwissClinicalTrialOrganisation/secuTrialR")
Recommended export options
While the package strives to allow loading of as many types of secuTrial data exports as possible, there are certain export options which are less likely to cause issues. If possible it is suggested to export data which adheres to a suggested option set. Thus, we suggest to work with exports which: - are zipped - are English - have reference values stored in a separate table - contain Add-IDs, centre information, structure information, form status, project setup - do NOT have the meta data duplicated into all tables - are UTF-8 encoded - are “CSV format” or “CSV format for MS Excel” - do NOT contain form data of hidden fields
If you use read_secuTrial() to read your export then it will inform you regarding deviations.
We also recommend using short names when exporting your data. Some users have reported issues importing data with long names, but the issues do not exist when using short names. That may (or may not) be related to upgarding SecuTrial.
Basic usage
An extensive applied manual/vignette is available here and probably the best place to get started.
Load the package
library(secuTrialR)
Load a dataset
export_location <- system.file("extdata", "sT_exports", "lnames",
"s_export_CSV-xls_CTU05_long_ref_miss_en_utf8.zip",
package = "secuTrialR")
ctu05 <- read_secuTrial(export_location)
## Read export successfully.
## The following export options deviate from the suggested specifications:
## Data from hidden fields is part of the export.
## Short names was not selected.
This will load all sheets from the export into an object of class secuTrialdata, which is basically a list. It will always contain export_details (which are parsed from the HTML ExportOptions file that secuTrial generates). By default, it will also contain all other files in the dataset. secuTrialR automatically strips file names of dates. The new file names can be seen via ctu05$export_options$data_names. The function also adds labels to variables and data.frames, converts categorical variables to factors and ensures that dates are Dates and date-times are POSIXct. read_secuTrial is a wrapper for the functions described below, so it is possible to achieve more flexibility by using the individual functions (if necessary). Individual tables can be extracted from the ctu05 object via tab <- ctu05$tab, where tab is the table of interest.
Wrapped functions
Load the dataset
# prepare path to example export
export_location <- system.file("extdata", "sT_exports", "BMD",
"s_export_CSV-xls_BMD_short_en_utf8.zip",
package = "secuTrialR")
# load all export data
bmd_export <- read_secuTrial_raw(data_dir = export_location)
# load a second dataset
export_location <- system.file("extdata", "sT_exports", "lnames",
"s_export_CSV-xls_CTU05_long_ref_miss_en_utf8.zip",
package = "secuTrialR")
ctu05_raw <- read_secuTrial_raw(export_location)
# View names of the bmd_export object
names(bmd_export)
## [1] "export_options" "fs" "cn" "ctr"
## [5] "is" "qs" "qac" "vp"
## [9] "vpfs" "atcn" "atcvp" "cts"
## [13] "bmd" "atbmd"
read_secuTrial_raw returns an object of class secuTrialdata, which is basically a list. It will always contain export_details (which are parsed from the HTML ExportOptions file that secuTrial generates). By default, it will also contain all other files in the dataset. secuTrialR automatically strips file names of dates. The new file names can be seen via bmd_export$export_options$data_names.
bmd_export is a list, with class secuTrialdata. To prevent it from printing all data to the console, a special print method returns some useful information about the objects within bmd_export instead. The information returned includes the original file name in the datafile, it’s name in the secuTrialdata object, together with the number of rows and columns and a column indicating whether the object is metadata or not:
bmd_export
## secuTrial data imported from:
## /Users/runner/work/_temp/Library/secuTrialR/extdata/sT_exports/BMD/s_export_CSV-xls_BMD_short_en_utf8.zip
## table nrow ncol meta original_name
## vp 1 10 TRUE vp.xls
## vpfs 1 2 TRUE vpfs.xls
## fs 1 7 TRUE fs.xls
## qs 1 7 TRUE qs.xls
## is 3 8 TRUE is.xls
## ctr 1 3 TRUE ctr.xls
## cn 113 13 TRUE cn.xls
## atcn 0 6 TRUE atcn.xls
## atcvp 0 11 TRUE atcvp.xls
## qac 0 10 TRUE qac.xls
## cts 0 8 TRUE cts.xls
## bmd 504 27 FALSE bmd.xls
## atbmd 0 28 FALSE atbmd.xls
Individual tables can be extracted from the bmd_export object via tab <- bmd_export$tab, where tab is the table of interest.
Variable labels
For creating tables, it is often useful to have access to variable labels. secuTrialR supports two main methods for handling them - a named list, or via variable attributes. The list approach works as follows.
labs <- labels_secuTrial(bmd_export)
# query the list with the variable name of interest
labs[["age"]]
## [1] "Age"
The attribute based approach adds labels as an attribute to a variable, which can then be accessed via label(var).
labelled <- label_secuTrial(bmd_export)
label(labelled$bmd$age)
## [1] "Age"
Labels can be added to new variables or changed via
label(labelled$bmd$age) <- "Age (years)"
label(labelled$bmd$age)
## [1] "Age (years)"
Where units have been defined in the SecuTrial database, they can be accessed or changed analogously (here, age had no unit assigned, but we can add one).
units(labelled$bmd$age)
## NULL
units(labelled$bmd$age) <- "years"
units(labelled$bmd$age)
## [1] "years"
There is a drawback to the attribute based approach - labels will not be propagated if variables are derived and may be lost if variables are edited.
Currently, label_secuTrial should be used prior to dates_secuTrial or factorize_secuTrial so that labels and units are propagated to factor and date variables.
Prepare factors
It is often useful to have categorical variables as factors (R knows how to handle factors). secuTrialR can prepare factors easily.
factors <- factorize_secuTrial(ctu05_raw)
This functions loops through each table of the dataset, creating new factor variables where necessary. The new variables are the same as the original but with .factor appended (i.e. a new variable called sex.factor would be added to the relevant form).
# original variable
str(factors$ctu05baseline$gender)
## int [1:17] 1 NA NA 2 1 2 1 NA NA 1 ...
# factor
str(factors$ctu05baseline$gender.factor)
## Factor w/ 2 levels "male","female": 1 NA NA 2 1 2 1 NA NA 1 ...
# cross tabulation
table(original = factors$ctu05baseline$gender, factor = factors$ctu05baseline$gender.factor)
## factor
## original male female
## 1 5 0
## 2 0 5
Prepare dates
Date(time)s are a very common data type. They cannot be easily used though in their export format. This is also easily rectified in secuTrialR:
dates <- dates_secuTrial(ctu05_raw)
Date variables are converted to Date class, and datetimes are converted to POSIXct class. Rather than overwriting the original variable, new variables are added with the new class. This is a safetly mechanism in case NAs are accidentally created.
dates$ctu05baseline[c(1, 7), c("aspirin_start", "aspirin_start.date",
"hiv_date", "hiv_date.datetime")]
## aspirin_start aspirin_start.date hiv_date hiv_date.datetime
## 1 NA <NA> 201903052356 2019-03-05 23:56:00
## 7 20060301 2006-03-01 NA <NA>
secuTrial exports containing date variables sometimes include incomplete dates. e.g. the day or the month may be missing. During date conversion (i.e. dates_secuTrial()) secuTrialR currently creates NAs from such incomplete date entries.
Incomplete dates are not approximated to exact dates, since this can lead to false conclusions and biases. Users are, however, informed about this behaviour with a warning(). Subsequent approximation of incomplete dates can be manually performed.
Recommended literature on incomplete dates/date imputation:
Dubois and Hebert 2001
Bowman 2006
Recommended approach if not using read_secuTrial
f <- "PATH_TO_FILE"
d <- read_secuTrial_raw(f)
l <- label_secuTrial(d)
fa <- factorize_secuTrial(l)
dat <- dates_secuTrial(fa)
# or, if you like pipes
library(magrittr)
f <- "PATH_TO_FILE"
d <- read_secuTrial_raw(f)
dat <- d %>%
label_secuTrial() %>%
factorize_secuTrial() %>%
dates_secuTrial()
Exploratory helpers
secuTrialR has a couple of functions to help get to grips with a secuTrial data export. They are intended to be used in an exploratory manner only.
as.data.frame
Working with a list can be tiresome so secuTrialR provides a as.data.frame method to save the data.frames in the list to an environment of your choice. As a demonstration, we’ll create a new environment (env) and create the data.frames in there. In practice, using .GlobalEnv would probably be more useful.
env <- new.env()
ls(env)
## character(0)
names(ctu05)
## [1] "export_options" "forms"
## [3] "casenodes" "centres"
## [5] "items" "questions"
## [7] "queries" "visitplan"
## [9] "visitplanforms" "atcasenodes"
## [11] "atcasevisitplans" "comments"
## [13] "miv" "cl"
## [15] "atmiv" "ctu05baseline"
## [17] "atmnpctu05baseline" "ctu05outcome"
## [19] "atmnpctu05outcome" "ctu05treatment"
## [21] "atmnpctu05treatment" "ctu05allmedi"
## [23] "atmnpctu05allmedi" "ctu05studyterminat"
## [25] "atmnpctu05studyterminat" "ctu05ae"
## [27] "atmnpctu05ae" "ctu05sae"
## [29] "atmnpctu05sae" "emnpctu05surgeries"
## [31] "atemnpctu05surgeries" "atadverseevents"
as.data.frame(ctu05, envir = env)
ls(env)
## [1] "atadverseevents" "atemnpctu05surgeries"
## [3] "atmiv" "atmnpctu05ae"
## [5] "atmnpctu05allmedi" "atmnpctu05baseline"
## [7] "atmnpctu05outcome" "atmnpctu05sae"
## [9] "atmnpctu05studyterminat" "atmnpctu05treatment"
## [11] "ctu05ae" "ctu05allmedi"
## [13] "ctu05baseline" "ctu05outcome"
## [15] "ctu05sae" "ctu05studyterminat"
## [17] "ctu05treatment" "emnpctu05surgeries"
There are also options for selecting specific forms (option data.frames), changing names based on a named vector (option data.frames) or regex (options regex and rep), and specifying whether metadata objects should be returned (option meta).
Recruitment over time
Recruitment is an important cornerstone for every clinical trial. secuTrialR allows for straigt forward visualizion of recuitment over time for a given export file.
# show plot
# note that there is no line for Universitätsspital
# Basel because only one participant is registered for this centre
plot_recruitment(ctu05, cex = 1.5, rm_regex = "\\(.*\\)$")
# return the plot data
plot_recruitment(ctu05, return_data = TRUE)
## [[1]]
## date centre_id pat_count centre_name
## 11 2018-05-01 441 1 Universitätsspital Basel (RPACK)
## 1 2019-04-01 462 2 Charité Berlin (RPACK)
## 2 2019-04-02 462 3 Charité Berlin (RPACK)
## 3 2019-04-03 462 4 Charité Berlin (RPACK)
## 4 2019-04-04 462 5 Charité Berlin (RPACK)
## 5 2019-04-05 462 6 Charité Berlin (RPACK)
## 6 2019-04-11 461 7 Inselspital Bern (RPACK)
## 7 2019-04-12 461 8 Inselspital Bern (RPACK)
## 8 2019-04-13 461 9 Inselspital Bern (RPACK)
## 9 2019-04-14 461 10 Inselspital Bern (RPACK)
## 10 2019-04-15 461 11 Inselspital Bern (RPACK)
##
## [[2]]
## date centre_id pat_count centre_name
## 1 2019-04-01 462 1 Charité Berlin (RPACK)
## 2 2019-04-02 462 2 Charité Berlin (RPACK)
## 3 2019-04-03 462 3 Charité Berlin (RPACK)
## 4 2019-04-04 462 4 Charité Berlin (RPACK)
## 5 2019-04-05 462 5 Charité Berlin (RPACK)
##
## [[3]]
## date centre_id pat_count centre_name
## 6 2019-04-11 461 1 Inselspital Bern (RPACK)
## 7 2019-04-12 461 2 Inselspital Bern (RPACK)
## 8 2019-04-13 461 3 Inselspital Bern (RPACK)
## 9 2019-04-14 461 4 Inselspital Bern (RPACK)
## 10 2019-04-15 461 5 Inselspital Bern (RPACK)
##
## [[4]]
## date centre_id pat_count centre_name
## 11 2018-05-01 441 1 Universitätsspital Basel (RPACK)
Furthermore, recruitment per year and center can be returned.
annual_recruitment(ctu05, rm_regex = "\\(.*\\)$")
## Center Total 2018 2019
## 1 All 11 1 10
## 2 Charité Berlin 5 0 5
## 3 Inselspital Bern 5 0 5
## 4 Universitätsspital Basel 1 1 0
Form status summary statistics
If you are not sure about how complete the data in you export is, it may be useful to get a quick overview of how well the forms have been filled.
count_summary <- form_status_summary(ctu05)
tail(count_summary)
## form_name partly_filled completely_filled empty with_warnings
## 5 ctu05allmedi 1 16 0 0
## 6 ctu05baseline 3 14 0 0
## 7 ctu05outcome 1 12 0 0
## 8 ctu05sae 0 2 0 0
## 9 ctu05studyterminat 0 10 0 0
## 10 ctu05treatment 0 11 0 0
## with_errors partly_filled.percent completely_filled.percent empty.percent
## 5 0 0.05882353 0.9411765 0
## 6 0 0.17647059 0.8235294 0
## 7 0 0.07692308 0.9230769 0
## 8 0 0.00000000 1.0000000 0
## 9 0 0.00000000 1.0000000 0
## 10 0 0.00000000 1.0000000 0
## with_warnings.percent with_errors.percent form_count
## 5 0 0 17
## 6 0 0 17
## 7 0 0 13
## 8 0 0 2
## 9 0 0 10
## 10 0 0 11
As you can see, the majority of forms has been completeley filled. None of the forms were saved empty, with warnings or with errors. For a more participant id centered statistic you can perform the following.
form_status_counts(ctu05)
This will give you a count based overview per participant id and form. Please note that both form_status_summary and form_status_counts only work with saved forms since unsaved form data is not available in secuTrial exports.
Visit plan
secuTrialR can provide a depiction of the visit structure, although only where the visit plan is fixed:
vs <- visit_structure(ctu05)
plot(vs)
Data dictionary
It can be difficult to find the variable you’re looking for. secuTrialR provides the dictionary_secuTrial function to help:
head(dictionary_secuTrial(ctu05))
## formtablename formname ffcolname itemtype
## 1 emnpctu05surgeries Surgeries surgery_organ Popup (Label Group)
## 2 emnpctu05surgeries Surgeries surgery_type Popup (Label Group)
## 3 mnpctu05ae Adverse Events ae_is_sae Horizontal Radiobutton
## 4 mnpctu05ae Adverse Events ae_end_time Checked Time (hh:mm)
## 5 mnpctu05ae Adverse Events ae_description Textarea 9,60
## 6 mnpctu05ae Adverse Events notes Textarea 9,60
## fflabel unit formfamily
## 1 Organ <NA> Subforms
## 2 Type <NA> Subforms
## 3 Is an SAE <NA> Adverse Events
## 4 Timepoints <NA> Adverse Events
## 5 Description of Adverse Event <NA> Adverse Events
## 6 Notes <NA> Adverse Events
Linking different forms
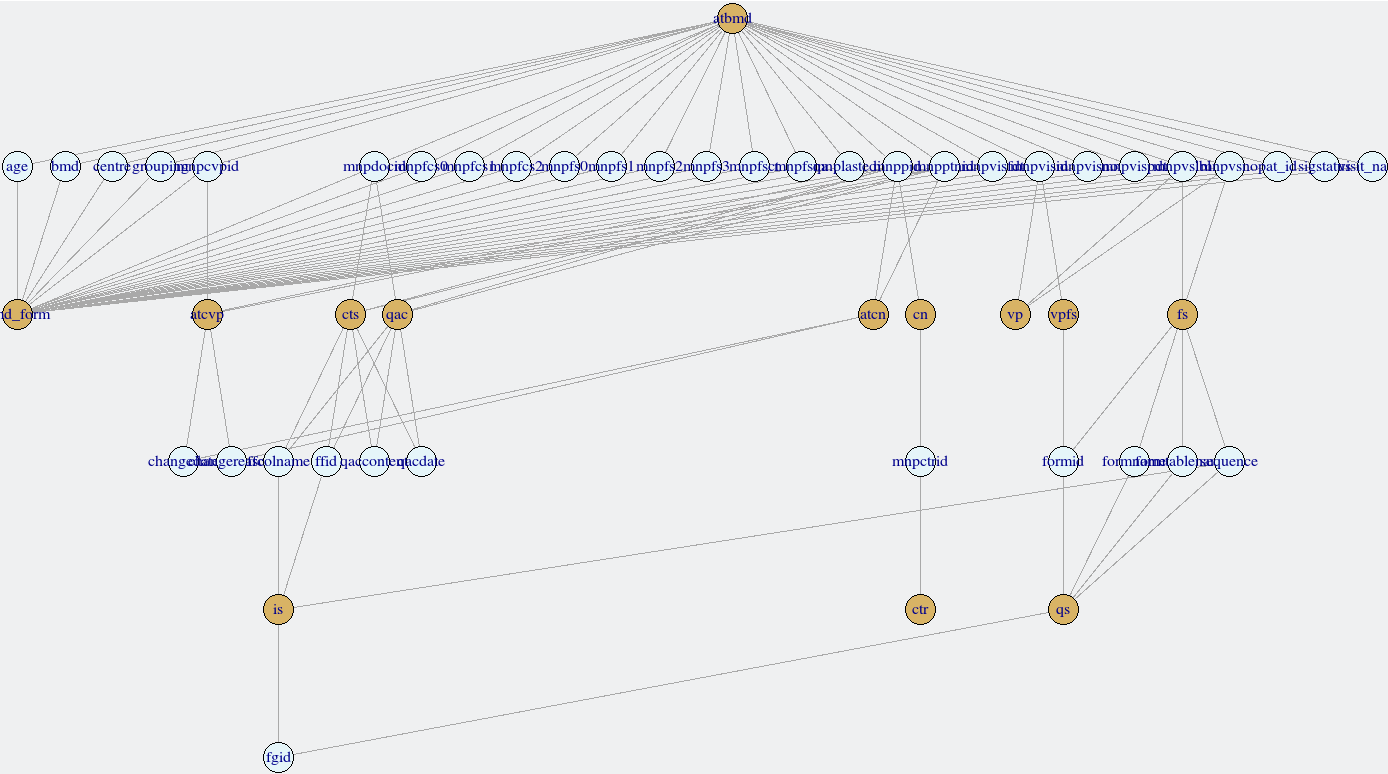
Linkages amongst forms can be explored with the links_secuTrial function. This relies on the igraph package to create a network. It is possible to interact with the network, e.g. move nodes around in order to read the labels better. The device ID is returned to the console, but can be ignored. Forms are plotted in deep yellow, variables in light blue.
links_secuTrial(bmd_export)
Sampling random participants
During study monitoring it is common practice to check random participants from a study database. These participants should be retrieved in a reproducible fashion. The below function allows this for a loaded secuTrial data export.
# retrieve at least 25 percent of participants recorded after March 18th 2019
# from the centres "Inselspital Bern" and "Charité Berlin"
return_random_participants(ctu05, percent = 0.25, seed = 1337, date = "2019-03-18",
centres = c("Inselspital Bern (RPACK)", "Charité Berlin (RPACK)"))
## $participants
## mnpaid centre mnpvisstartdate
## 2 RPACK-INS-012 Inselspital Bern (RPACK) 2019-04-12
## 4 RPACK-INS-014 Inselspital Bern (RPACK) 2019-04-14
## 5 RPACK-CBE-005 Charité Berlin (RPACK) 2019-04-05
## 3 RPACK-CBE-003 Charité Berlin (RPACK) 2019-04-03
##
## $rng_config
## [1] "Mersenne-Twister" "Inversion" "Rejection"
For contributors
Testing with devtools
# run tests
devtools::test("secuTrialR")
# spell check -> will contain some technical terms beyond the below list which is fine
ignore_words <- c("AdminTool", "allforms", "casenodes", "CDMS", "codebook",
"codebooks", "datetime" ,"dir" ,"Hmisc" ,"igraph",
"labelled", "mnp", "savedforms", "secutrial", "secuTrial",
"secuTrialdata", "tcltk", "tibble")
devtools::spell_check("secuTrialR", ignore = ignore_words)
Linting with lintr
# lint the package -> should be clean
library(lintr)
lint_package("secuTrialR", linters = with_defaults(camel_case_linter = NULL,
object_usage_linter = NULL,
line_length_linter(125)))
Building the vignette
library(rmarkdown)
render("vignettes/secuTrialR-package-vignette.Rmd",
output_format=c("pdf_document"))
Generating the README file
The README file is automatically generated on GitHub via a GitHub action.
Handling dependencies
Dependencies to other R packages are to be declared in the DESCRIPTION file under Imports: and in the specific roxygen2 documentation of the functions relying on the dependency. It is suggested to be as explicit as possible. i.e. Just import functions that are needed and not entire packages.
Example to import str_matchstr_lengthstr_wrap from the stringr package (see read_secuTrial_raw.R):
#' @importFrom stringr str_match str_length str_wrap
Preparing a release on CRAN
# build the package archive
R CMD build secuTrialR
# check the archive (should return "Status: OK", no WARNINGs, no NOTEs)
# in this example for version 0.9.0
R CMD check secuTrialR_0.9.0.tar.gz
Versioning and releases
The version number is made up of three digits. The first digit is reserved for major releases which may break backwards compatibility. The second and third digits are used for medium and minor changes respectively. Versions released on CRAN will be tagged and saved as releases on GitHub. The version released on CRAN is regarded as the stable version while the master branch on GitHub is regarded as the current development version.
Release checklist
Compile/Update: * README.Rmd * vignette * pkgdown page * NEWS.md
Guidelines for contributors
Requests for new features and bug fixes should first be documented as an Issue on GitHub. Subsequently, in order to contribute to this R package you should fork the main repository. After you have made your changes please run the tests and lint your code as indicated above. Please also increment the version number and recompile the README.md to increment the dev-version badge (requires installing the package after editing the DESCRIPTION file). If all tests pass and linting confirms that your coding style conforms you can send a pull request (PR). Changes should also be mentioned in the NEWS file. The PR should have a description to help the reviewer understand what has been added/changed. New functionalities must be thoroughly documented, have examples and should be accompanied by at least one test to ensure long term robustness. The PR will only be reviewed if all travis checks are successful. The person sending the PR should not be the one merging it.
A depiction of the core functionalities for loading can be found here.
Citation 
If you use and benefit from secuTrialR in your work please cite it as:
Wright et al., (2020). secuTrialR: Seamless interaction with clinical trial databases in R. Journal of Open Source Software, 5(55), 2816, https://doi.org/10.21105/joss.02816
