Metrics for Assessing Segmentation Accuracy for Geospatial Data.
segmetric 
Segmentation Assessment Metrics (segmetric)
The segmetric is an open source package that provides a set of metrics for analyzing and evaluating geospatial segmentations. It implements 28 supervised metrics used in literature for spatial segmentation assessment (see References below).
Installation
# install via CRAN
install.packages("segmetric")
Development version
To install the development version of segmetric, run the following commands:
# load necessary libraries
library(devtools)
install_github("michellepicoli/segmetric")
Usage
Spatial datasets can be loaded using sf objects. To create a segmetric object, use function sm_read():
library(segmetric)
# load example datasets
data("sample_ref_sf", package = "segmetric")
data("sample_seg_sf", package = "segmetric")
# create segmetric object
m <- sm_read(ref_sf = sample_ref_sf, seg_sf = sample_seg_sf)
Plot your data using plot() command:
plot(m)
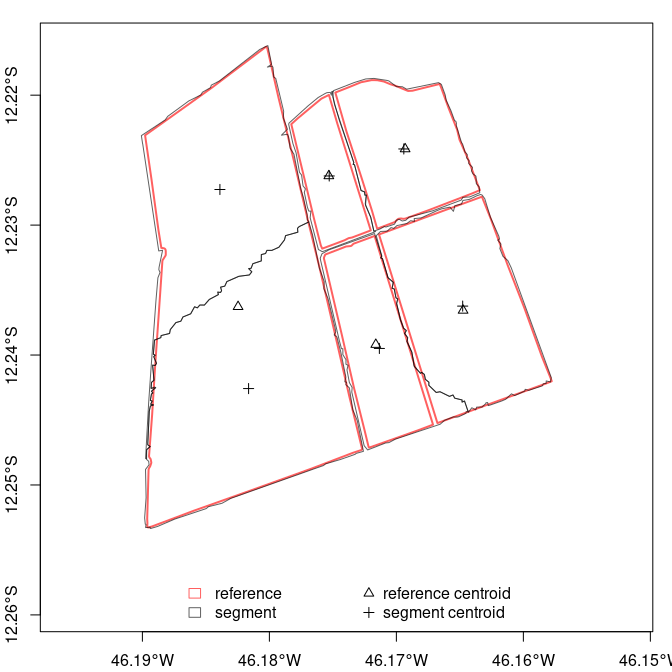
Segmentation metrics can be computed by function sm_compute(). Use summary() to obtain an overall metric (mean or weighted mean).
# compute AFI metric and summarize it
sm_compute(m, "AFI") %>% summary()
#> [1] -0.007097452
Make multiple calls to compute more other metrics:
# compute OS1, F_measure, and US2 metrics
m <-
sm_compute(m, "OS1") %>%
sm_compute("F_measure") %>%
sm_compute("US2")
# summarize them
summary(m)
#> OS1 F_measure US2
#> 0.17341468 0.84728616 0.08617454
To see all supported metrics, type ?metric_functions or run:
# list all supported metrics
sm_list_metrics()
#> [1] "AFI" "D_index" "Dice" "E" "ED3" "F_measure"
#> [7] "Fitness" "IoU" "M" "OI2" "OMerging" "OS1"
#> [13] "OS2" "OS3" "PI" "precision" "qLoc" "QR"
#> [19] "RAsub" "RAsuper" "recall" "RPsub" "RPsuper" "SimSize"
#> [25] "UMerging" "US1" "US2" "US3"
Getting Help
A detailed documentation with examples on how to use each function inside segmetric package can be obtained by typing ?segmetric in R console.
How to contribute?
The segmetric package was implemented based on an extensible architecture. Feel free to contribute by implementing new metrics functions.
- Make a project fork.
- Edit file
R/metric-funs.Rimplementing the new metric. - Register your metric in
.db_registry()function atR/db.Rfile usingsm_reg_metric(). - Make a Pull Request on the branch dev.
Acknowledgements
This research was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (Grant agreement No 677140 MIDLAND).
References
- Carleer, A.P., Debeir, O., Wolff, E., 2005. Assessment of very high spatial resolution satellite image segmentations. Photogramm. Eng. Remote. Sens. 71, 1285-1294. http://dx.doi.org/10.14358/PERS.71.11.1285.
- Clinton, N., Holt, A., Scarborough, J., Yan, L., Gong, P., 2010. Accuracy assessment measures for object-based image segmentation goodness. Photogramm. Eng. Remote. Sens. 76, pp. 289-299.
- Costa, G.A.O.P., Feitosa, R.Q., Cazes, T.B., Feijo, B., 2008. Genetic adaptation of segmentation parameters. In: Blaschke, T., Lang, S., Hay, G.J. (Eds.), Object-based Image Analysis. Springer Berlin Heidelberg, Berlin, Heidelberg, pp. 679-695. http://dx.doi.org/10.1007/978-3-540-77058-9_37.
- Dice, L.R., 1945. Measures of the amount of ecologic association between species. Ecology, 26(3), pp.297-302.
- Feitosa, R.Q., Ferreira, R.S., Almeida, C.M., Camargo, F.F., Costa, G.A.O.P., 2010. Similarity metrics for genetic adaptation of segmentation parameters. In: 3rd International Conference on Geographic Object-Based Image Analysis (GEOBIA 2010). The International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences, Ghent.
- Jaccard, P., 1912. The distribution of the flora in the alpine zone.
- New phytologist, 11(2), pp.37-50. http://dx.doi.org/10.1111/j.1469-8137.1912.tb05611.x
- Janssen, L.L.F., Molenaar, M., 1995. Terrain objects, their dynamics and their monitoring by the integration of GIS and remote sensing. IEEE Trans. Geosci. Remote Sens. 33, pp. 749-758. http://dx.doi.org/10.1109/36.387590.
- Levine, M.D., Nazif, A.M., 1982. An experimental rule based system for testing low level segmentation strategies. In: Preston, K., Uhr, L. (Eds.), Multicomputers and Image Processing: Algorithms and Programs. Academic Press, New York, pp. 149-160.
- Lucieer, A., Stein, A., 2002. Existential uncertainty of spatial objects segmented from satellite sensor imagery. Geosci. Remote. Sens. IEEE Trans. 40, pp. 2518-2521. http://dx.doi.org/10.1109/TGRS.2002.805072.
- Moller, M., Lymburner, L., Volk, M., 2007. The comparison index: a tool for assessing the accuracy of image segmentation. Int. J. Appl. Earth Obs. Geoinf. 9, pp. 311-321. http://dx.doi.org/10.1016/j.jag.2006.10.002.
- Persello, C., Bruzzone, L., 2010. A novel protocol for accuracy assessment in classification of very high resolution images. IEEE Trans. Geosci. Remote Sens. 48, pp. 1232-1244. http://dx.doi.org/10.1109/TGRS.2009.2029570.
- Rezatofighi, H., Tsoi, N., Gwak, J., Sadeghian, A., Reid, I., Savarese, S.,
- In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 658-666.
- Van Coillie, F.M.B., Verbeke, L.P.C., De Wulf, R.R., 2008. Semi-automated forest stand delineation using wavelet based segmentation of very high resolution optical imagery. In: Object-Based Image Analysis: Spatial Concepts for Knowledge-Driven Remote Sensing Applications, pp. 237-256. http://dx.doi.org/10.1007/978-3-540-77058-9_13.
- Van Rijsbergen, C.J., 1979. Information Retrieval. Butterworth-Heinemann, London.
- Weidner, U., 2008. Contribution to the assessment of segmentation quality for remote sensing applications. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 37, pp. 479-484.
- Yang, J., Li, P., He, Y., 2014. A multi-band approach to unsupervised scale parameter selection for multi-scale image segmentation. ISPRS J. Photogramm. Remote Sens. 94, pp. 13-24. http://dx.doi.org/10.1016/j.isprsjprs.2014.04.008.
- Yang, J., He, Y., Caspersen, J. P., Jones, T. A., 2017. Delineating Individual Tree Crowns in an Uneven-Aged, Mixed Broadleaf Forest Using Multispectral Watershed Segmentation and Multiscale Fitting. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens., 10(4), pp. 1390-1401. http://dx.doi.org/10.1109/JSTARS.2016.2638822.
- Zhan, Q., Molenaar, M., Tempfli, K., Shi, W., 2005. Quality assessment for geo‐spatial objects derived from remotely sensed data. International Journal of Remote Sensing, 26(14), pp.2953-2974. http://dx.doi.org/10.1080/01431160500057764.
- Zhang, X., Feng, X., Xiao, P., He, G., Zhu, L., 2015a. Segmentation quality evaluation using region-based precision and recall measures for remote sensing images. ISPRS J. Photogramm. Remote Sens. 102, pp. 73-84. http://dx.doi.org/10.1016/j.isprsjprs.2015.01.009.

