Modular Cohort-Building Framework for Analytical Dashboards.
shinyCohortBuilder 
Move your cohortBuilder workflow to Shiny.
Installation
# CRAN version
install.packages("shinyCohortBuilder")
# Latest development version
remotes::install_github("https://github.com/r-world-devs/shinyCohortBuilder")
Overview
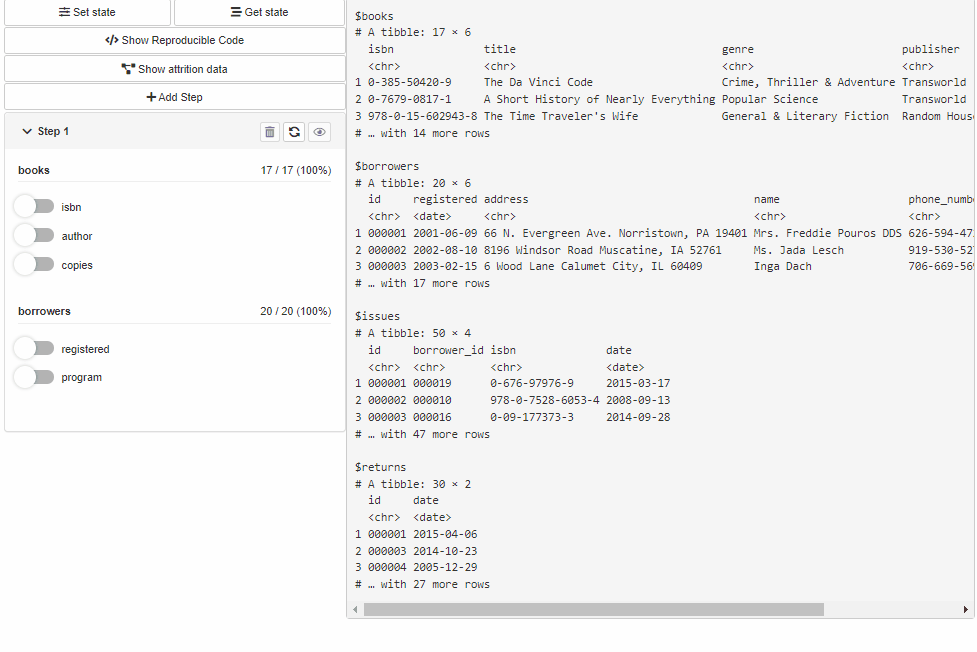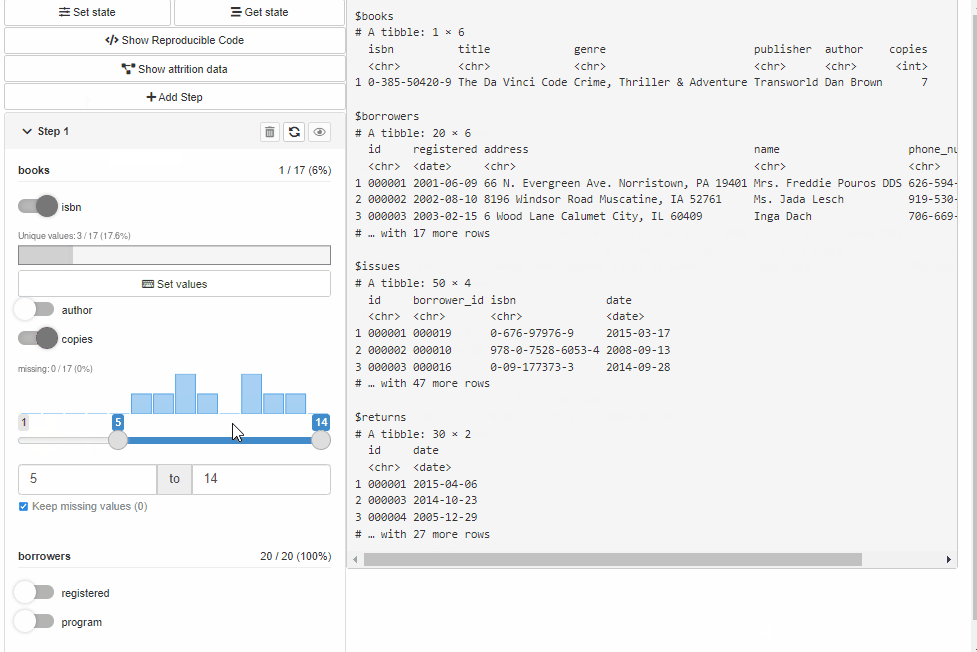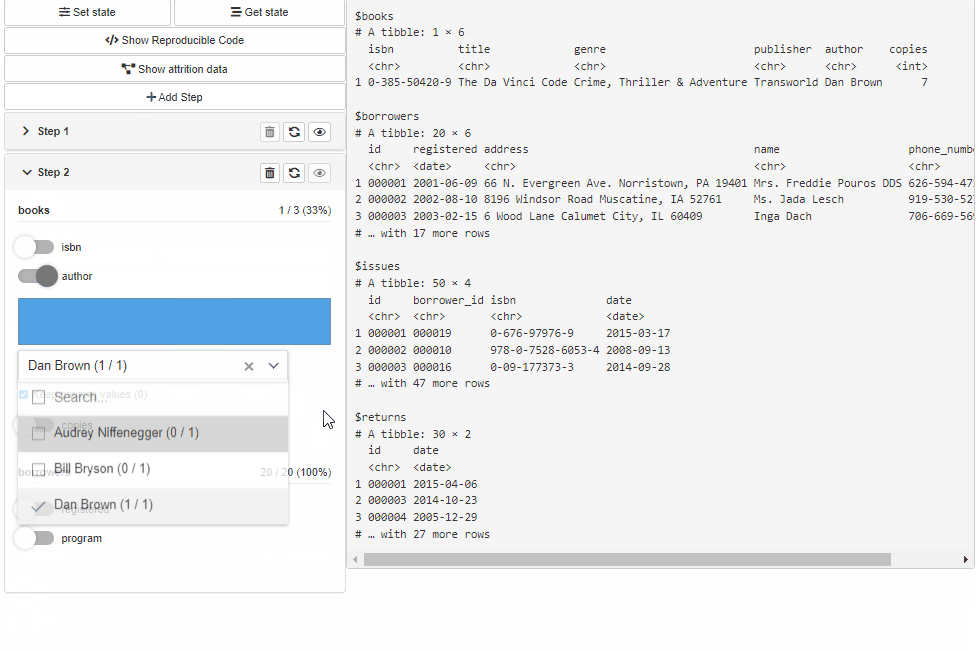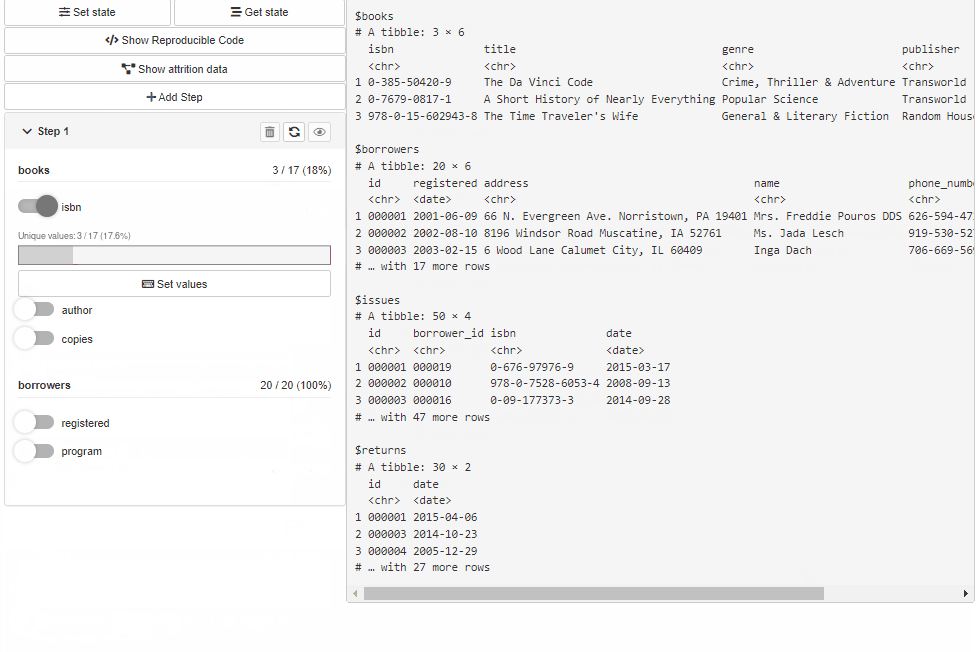
With shinyCohortBuilder you can use cohortBuilder features within your shiny application.
Configure Source and Cohort filters with cohortBuilder (set value/range to NA to select all the options / the whole range, and active = FALSE to collapse filter in GUI):
librarian_source <- set_source(as.tblist(librarian))
librarian_cohort <- cohort(
librarian_source,
filter(
"discrete", id = "author", dataset = "books",
variable = "author", value = "Dan Brown",
active = FALSE
),
filter(
"range", id = "copies", dataset = "books",
variable = "copies", range = c(5, 10),
active = FALSE
),
filter(
"date_range", id = "registered", dataset = "borrowers",
variable = "registered", range = c(as.Date("2010-01-01"), Inf),
active = FALSE
)
)
And apply in your application with cb_ui and cb_server:
library(shiny)
ui <- fluidPage(
sidebarLayout(
sidebarPanel(
cb_ui("librarian")
),
mainPanel()
)
)
server <- function(input, output, session) {
cb_server("librarian", librarian_cohort)
}
shinyApp(ui, server)
You may listen to cohort data changes with input[[<cohort-id>-data-updated]]:
library(shiny)
ui <- fluidPage(
sidebarLayout(
sidebarPanel(
cb_ui("librarian")
),
mainPanel(
verbatimTextOutput("cohort_data")
)
)
)
server <- function(input, output, session) {
cb_server("librarian", librarian_cohort)
output$cohort_data <- renderPrint({
input[["librarian-data-updated"]]
get_data(librarian_cohort)
})
}
shinyApp(ui, server)
Or run filtering panel locally what just makes your work with cohortBuilder easier:
gui(librarian_cohort)
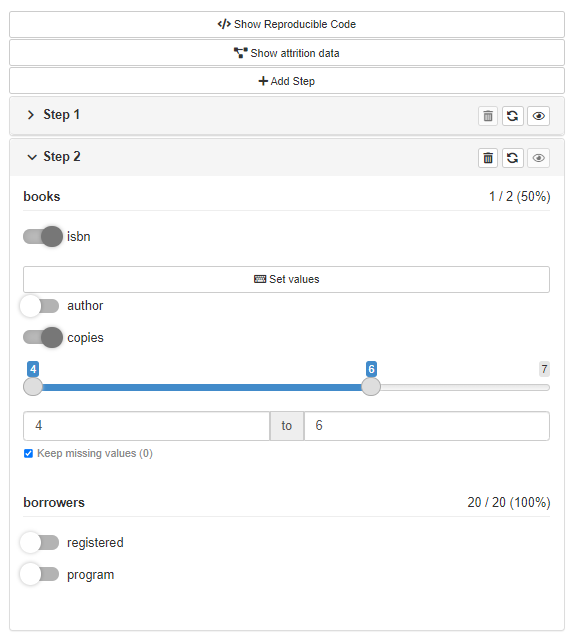
If you’re interested in more features of shinyCohortBuilder please visit the package website.
Acknowledgement
Special thanks to:
- Kamil Wais for highlighting the need for the package and its relevance to real-world applications.
- Adam Foryś for technical support, numerous suggestions for the current and future implementation of the package.
- Paweł Kawski for indication of initial assumptions about the package based on real-world medical data.
Getting help
In a case you found any bugs, have feature request or general question please file an issue at the package Github. You may also contact the package author directly via email at [email protected].

