Visual Documentation for Data Preprocessing.

smallsets: Visual Documentation for Data Preprocessing in R and Python
smallsets website: lydialucchesi.github.io/smallsets/
Do you use R or Python to preprocess datasets for analyses? smallsets is an R package (https://CRAN.R-project.org/package=smallsets) that transforms the preprocessing code in your R, R Markdown, Python, or Jupyter Notebook file into a Smallset Timeline. A Smallset Timeline is a static, compact visualisation composed of small data snapshots of different preprocessing steps. A full description of the Smallset Timeline can be found in the paper Smallset Timelines: A Visual Representation of Data Preprocessing Decisions in the proceedings of ACM FAccT ’22. A short (3 min) and long (15 min) YouTube video provide an introduction to the project.
The smallsets user guide is available here and in the package in vignette("smallsets"). If you have questions or would like help building a Smallset Timeline, please email Lydia.
Download the smallsets cheatsheet (1-page PDF)
Install from CRAN
install.packages("smallsets")
Quick start example
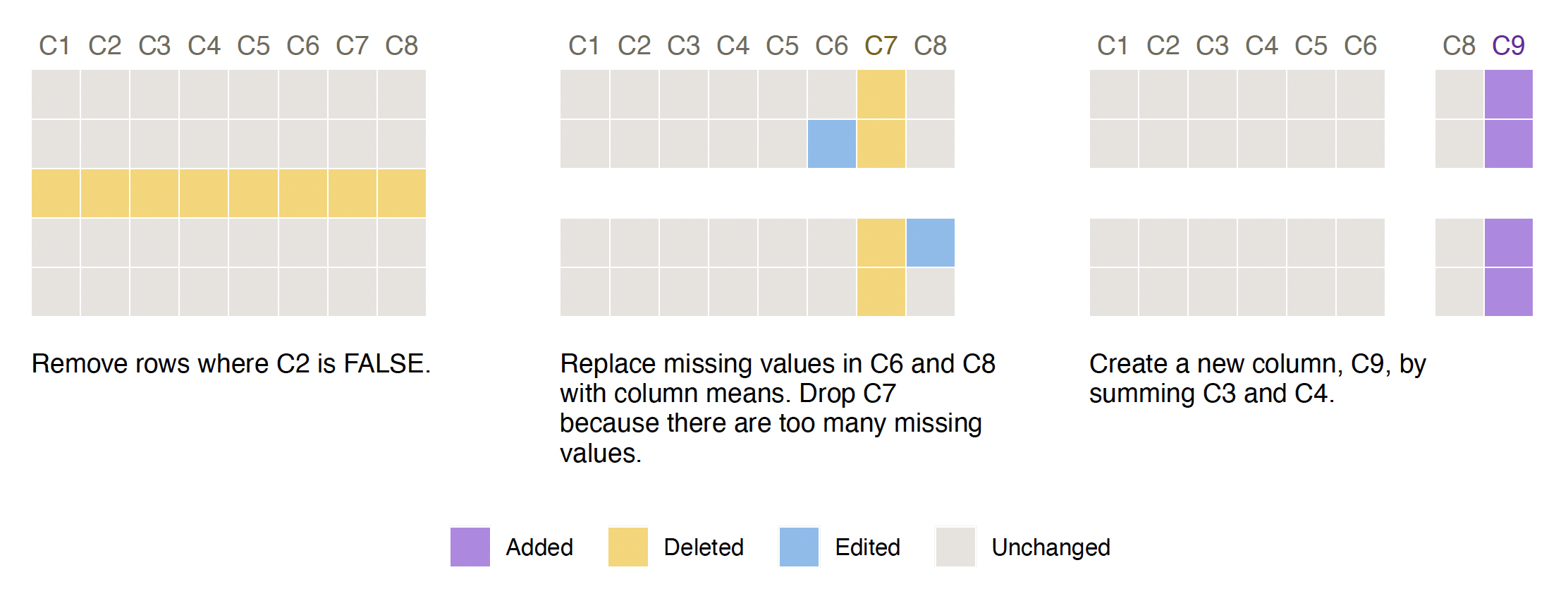
Run this snippet of code to build your first Smallset Timeline! It’s based on the synthetic dataset s_data, with 100 observations and eight variables (C1-C8), and the preprocessing script s_data_preprocess.R, discussed in the following section.
library(smallsets)
set.seed(145)
Smallset_Timeline(data = s_data,
code = system.file("s_data_preprocess.R", package = "smallsets"))
Structured comments
The Smallset Timeline above is based on the R preprocessing script below, s_data_preprocess.R. Structured comments were added to it, informing smallsets what to do.
# smallsets snap s_data caption[Remove rows where C2 is FALSE.]caption
s_data <- s_data[s_data$C2 == TRUE, ]
# smallsets snap +2 s_data caption[Replace missing values in C6 and C8 with
# column means. Drop C7 because there are too many missing values.]caption
s_data$C6[is.na(s_data$C6)] <- mean(s_data$C6, na.rm = TRUE)
s_data$C8[is.na(s_data$C8)] <- mean(s_data$C8, na.rm = TRUE)
s_data$C7 <- NULL
# smallsets snap +1 s_data caption[Create a new column, C9, by summing C3 and
# C4.]caption
s_data$C9 <- s_data$C3 + s_data$C4
Citing smallsets
If you use the smallsets software, please cite the Smallset Timeline paper.
Lydia R. Lucchesi, Petra M. Kuhnert, Jenny L. Davis, and Lexing Xie. 2022. Smallset Timelines: A Visual Representation of Data Preprocessing Decisions. In 2022 ACM Conference on Fairness, Accountability, and Transparency (FAccT ’22). Association for Computing Machinery, New York, NY, USA, 1136–1153. https://doi.org/10.1145/3531146.3533175
@inproceedings{SmallsetTimelines,
author = {Lucchesi, Lydia R. and Kuhnert, Petra M. and Davis, Jenny L. and Xie, Lexing},
title = {Smallset Timelines: A Visual Representation of Data Preprocessing Decisions},
year = {2022},
isbn = {9781450393522},
publisher = {Association for Computing Machinery},
address = {New York, NY, USA},
url = {https://doi.org/10.1145/3531146.3533175},
doi = {10.1145/3531146.3533175},
location = {Seoul, Republic of Korea},
series = {FAccT '22}
}