Conducting and Visualizing Specification Curve Analyses.
specr
Conducting and Visualizing Specification Curve Analyses
News
20 January 2022: specr version 1.0.0 is now available via github. This is a major update with several new features and functions. Note: it introduces a new framework for conduction specification curve analyses compared to earlier versions (see version history for more details).
4 December 2020: specr development version 0.2.2 is available via github. Mostly minor updates and bug fixes.
25 May 2020: specr version 0.2.1 has been released on CRAN.
What is specr?
The goal of specr is to facilitate specification curve analyses (Simonsohn, Simmons & Nelson, 2020; also known as multiverse analyses, see Steegen, Tuerlinckx, Gelman & Vanpaemel, 2016). The package can be used to investigate how different (theoretically plausible) analytical choices affect outcome statistics within the universe of one single data set. It provides functions to setup, run, evaluate, and plot the multiverse of specifications. A simple example of how to use specr is provided below. For more information about the various functions and specific vignettes and use cases, visit the documentation.
Disclaimer
We do see a lot of value in investigating how analytical choices affect a statistical outcome of interest. However, we strongly caution against using specr as a tool to somehow arrive at a better estimate. Running a specification curve analysis does not make your findings any more reliable, valid or generalizable than a single analysis. The method is only meant to inform about the effects of analytical choices on results, and not a better way to estimate a correlation or effect.
Installation
Install specr from CRAN:
install.packages("specr") # version 0.2.1
Or install the most recent development version from GitHub with:
# install.packages("devtools")
devtools::install_github("masurp/specr") # version 1.0.0
Usage
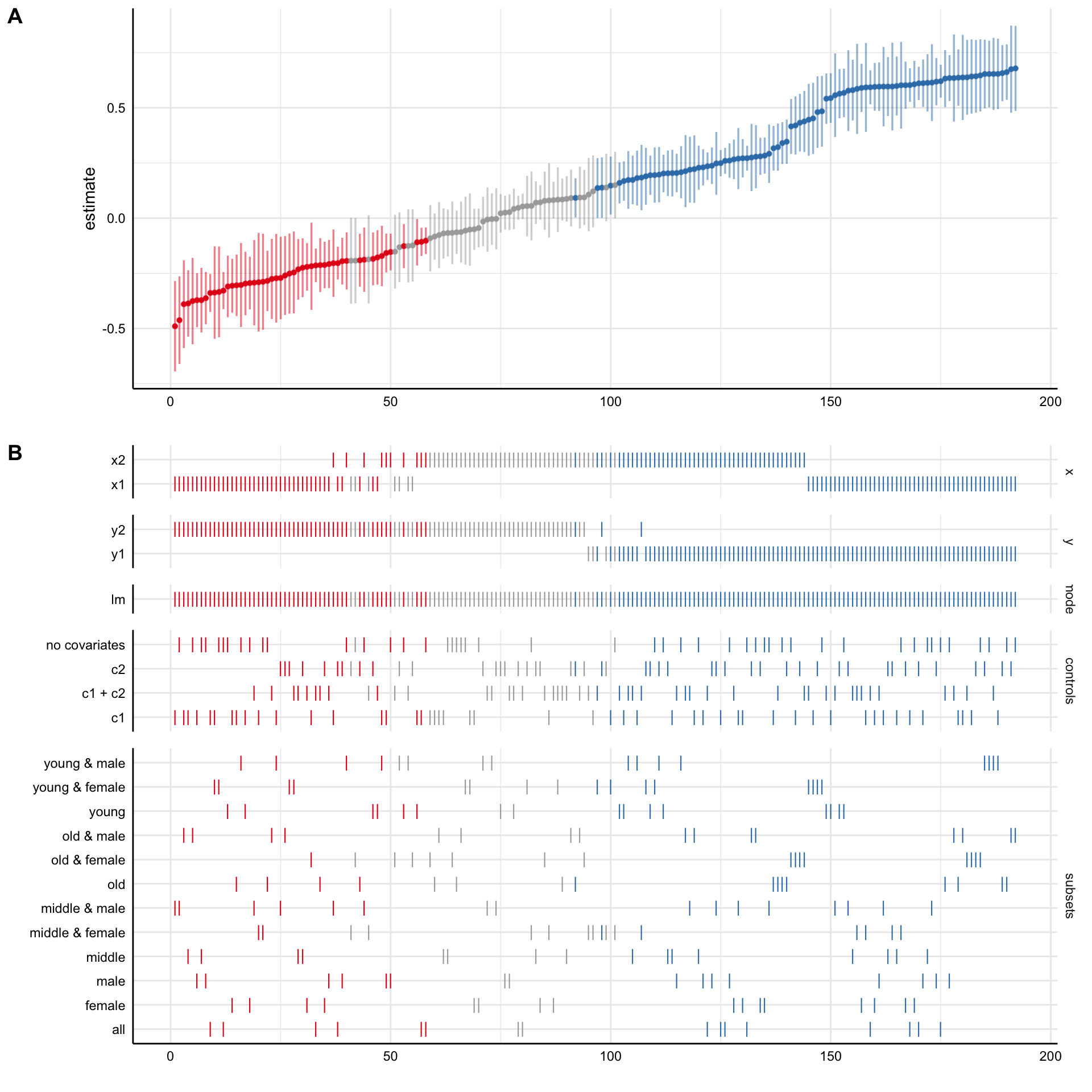
Using specr is comparatively simple. The two main function are setup(), in which analytic choices are specified as arguments, and specr(), which fits the models across all specifications. The latter creates a class called “specr.object”, which can be summarized and plotted with generic function such as summary or plot.
# Load package ----
library(specr)
# Setup Specifications ----
specs <- setup(data = example_data,
y = c("y1", "y2"),
x = c("x1", "x2"),
model = c("lm"),
controls = c("c1", "c2"),
subsets = list(group1 = unique(example_data$group1),
group2 = unique(example_data$group2)))
# Run Specification Curve Analysis ----
results <- specr(specs)
# Plot Specification Curve ----
plot(results)
How to cite this package
citation("specr")
#>
#> To cite 'specr' in publications use:
#>
#> Masur, Philipp K. & Scharkow, M. (2020). specr: Conducting and
#> Visualizing Specification Curve Analyses. Available from
#> https://CRAN.R-project.org/package=specr.
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Misc{,
#> title = {specr: Conducting and Visualizing Specification Curve Analyses (Version 1.0.0)},
#> author = {Philipp K. Masur and Michael Scharkow},
#> year = {2020},
#> url = {https://CRAN.R-project.org/package=specr},
#> }
References
Simonsohn, U., Simmons, J.P. & Nelson, L.D. (2020). Specification curve analysis. Nature Human Behaviour, 4, 1208–1214. https://doi.org/10.1038/s41562-020-0912-z
Steegen, S., Tuerlinckx, F., Gelman, A., & Vanpaemel, W. (2016). Increasing Transparency Through a Multiverse Analysis. Perspectives on Psychological Science, 11(5), 702-712. https://doi.org/10.1177/1745691616658637
Papers that used ‘specr’
If you have published a paper in which you used specr and you would like to be included in the following list, please send an email to Philipp.
Akaliyski, P., Minkov, M., Li, J., Bond, M. H., & Gehring, S. (2022). The weight of culture: Societal individualism and flexibility explain large global variations in obesity. Social Science & Medicine, 307. https://doi.org/10.1016/j.socscimed.2022.115167
Ballou, N., & van Rooij, A. J. (2021). The relationship between mental well-being and dysregulated gaming: a specification curve analysis of core and peripheral criteria in five gaming disorder scales. The Royal Society Open Science.https://doi.org/10.1098/rsos.201385
Ballou, N., & Zendle, D. (2022). “Clinically significant distress” in internet gaming disorder: An individual participant meta-analysis. Computers in Human Behavior, 129. https://doi.org/10.1016/j.chb.2021.107140
Burton, J.W., Cruz, N. & Hahn, U. (2021). Reconsidering evidence of moral contagion in online social networks. Nature Human Behaviour.https://doi.org/10.1038/s41562-021-01133-5
Cosme, D., & Lopez, R. B. (2020, March 7). Neural indicators of food cue reactivity, regulation, and valuation and their associations with body composition and daily eating behavior. https://doi.org/10.1093/scan/nsaa155
Del Giudice, M., & Gangestad, S. W. (2021). A Traveler’s Guide to the Multiverse: Promises, Pitfalls, and a Framework for the Evaluation of Analytic Decisions. Advances in Methods and Practices in Psychological Science.https://doi.org/10.1177/2515245920954925
Henson, P., Rodriguez-Villa, E., Torous, J. (2021). Investigating Associations Between Screen Time and Symptomatology in Individuals With Serious Mental Illness: Longitudinal Observational Study Journal of Medical Internet Research, 23(3), e23144. https://doi.org/10.2196/23144
Huang, S., Lai, X., Zhao, X., Dai, X., Yao, Y., Zhang, C., & Wang, Y., (2022). Beyond screen time: Exploring associations between types of smartphone use content and adolescents’ social relationships. International Journal of Environmental Research and Public Health, 19, 8940. https://doi.org/10.3390/ijerph19158940
Kritzler, S., & Luhmann, M. (2021, March 25). Be Yourself and Behave Appropriately: Exploring Associations Between Incongruent Personality States and Positive Affect, Tiredness, and Cognitive Performance. https://doi.org/10.31234/osf.io/9utyj
Masur, P. K. (2021). Understanding the Effects of Conceptual and Analytical Choices on ‘Finding’ the Privacy Paradox: A Specification Curve Analysis of Large-Scale Survey Data. Information, Communication & Society.https://doi.org/10.1080/1369118X.2021.1963460
Rauvola, R. S., & Rudolph, C. W. (2023). Worker aging, control, and well-being: A specification curve analysis. Acta Psychologica, 233, 103833.
Yuan, Q., Li, H., Du, B., Dang, Q., Chang, Q., Zhang, Z., … & Guo, T. (2023). The cerebellum and cognition: further evidence for its role in language control. Cerebral Cortex, 33(1), 35-49. https://doi.org/10.1093/cercor/bhac051