Description
Longitudinal Regression Trees and Forests.
Description
Builds regression trees and random forests for longitudinal or functional data using a spline projection method. Implements and extends the work of Yu and Lambert (1999) <doi:10.1080/10618600.1999.10474847>. This method allows trees and forests to be built while considering either level and shape or only shape of response trajectories.
README.md
splinetree
This package allows users to create, visualize, and evaluate regression trees and random forests for longitudinal or functional data through a spline projection method first suggested by Yu and Lambert (1999).
Installation
You can install splinetree from CRAN or from github with:
# install.packages("devtools")
devtools::install_github("anna-neufeld/splinetree")
Usage
Detailed information on using this package can be found in the package vignettes. The package vignettes can be accessed with:
browseVignettes(package='splinetree')
The vignettes are also available on the package website, https://anna-neufeld.github.io/splinetree/reference/index.html.
Example Tree
library(splinetree)
#> Loading required package: rpart
#> Loading required package: nlme
#> Loading required package: splines
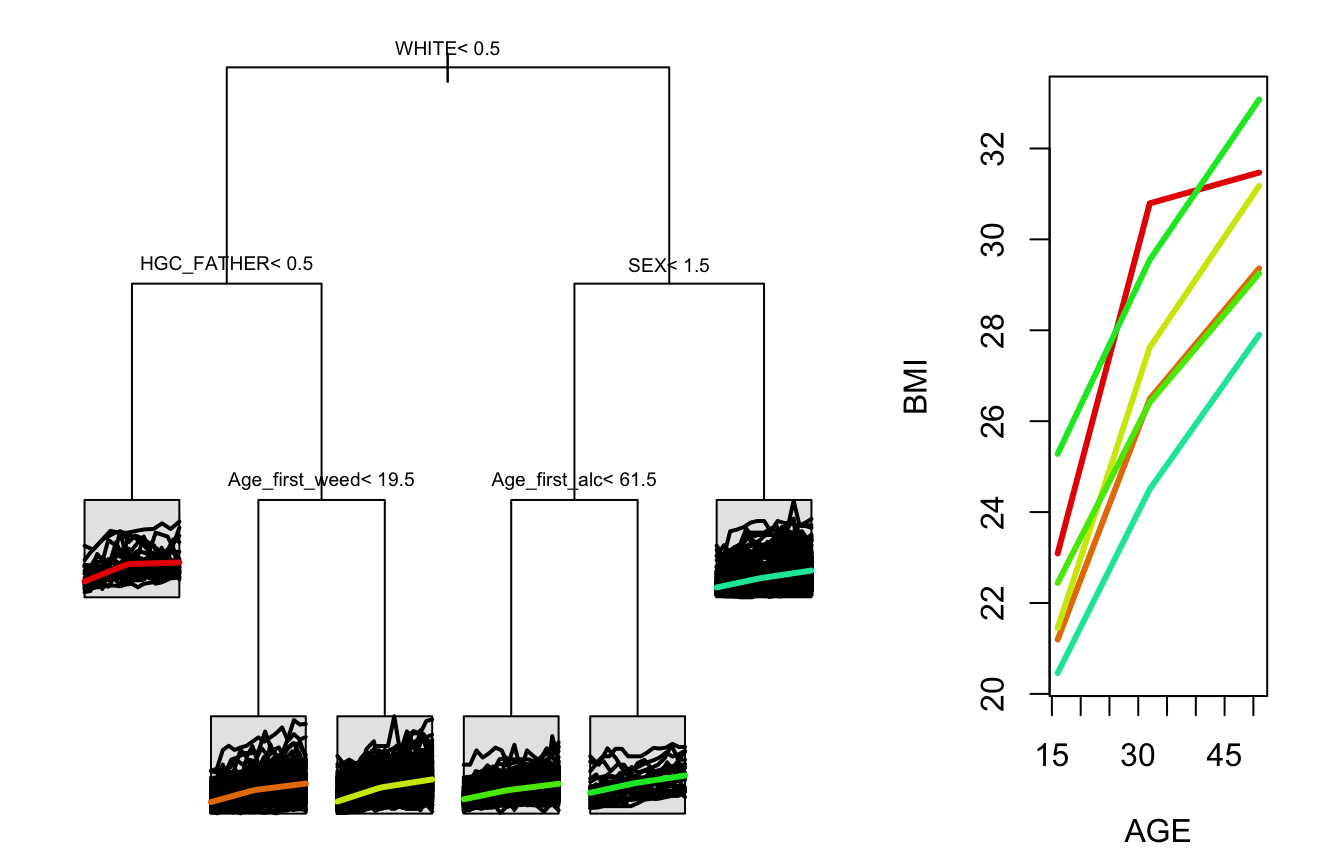
tree1 <- splineTree(~HISP+WHITE+BLACK+HGC_MOTHER+HGC_FATHER+SEX+Num_sibs,
BMI ~ AGE, "ID", nlsySample, degree = 1, df=2, intercept = FALSE, cp = 0.005)
stPrint(tree1)
#> n= 1000,
#>
#> node), split, n , coefficients
#> * denotes terminal node
#>
#> 1) root, 1000, (4.961796, 8.091247)
#> 2) WHITE< 0.5, 505, (5.882807, 9.006190)*
#> 3) WHITE>=0.5, 495, (4.022179, 7.157821)
#> 6) HGC_FATHER< 8.5, 78, (5.198284, 8.642817)*
#> 7) HGC_FATHER>=8.5, 417, (3.802188, 6.880053)*
stPlot(tree)
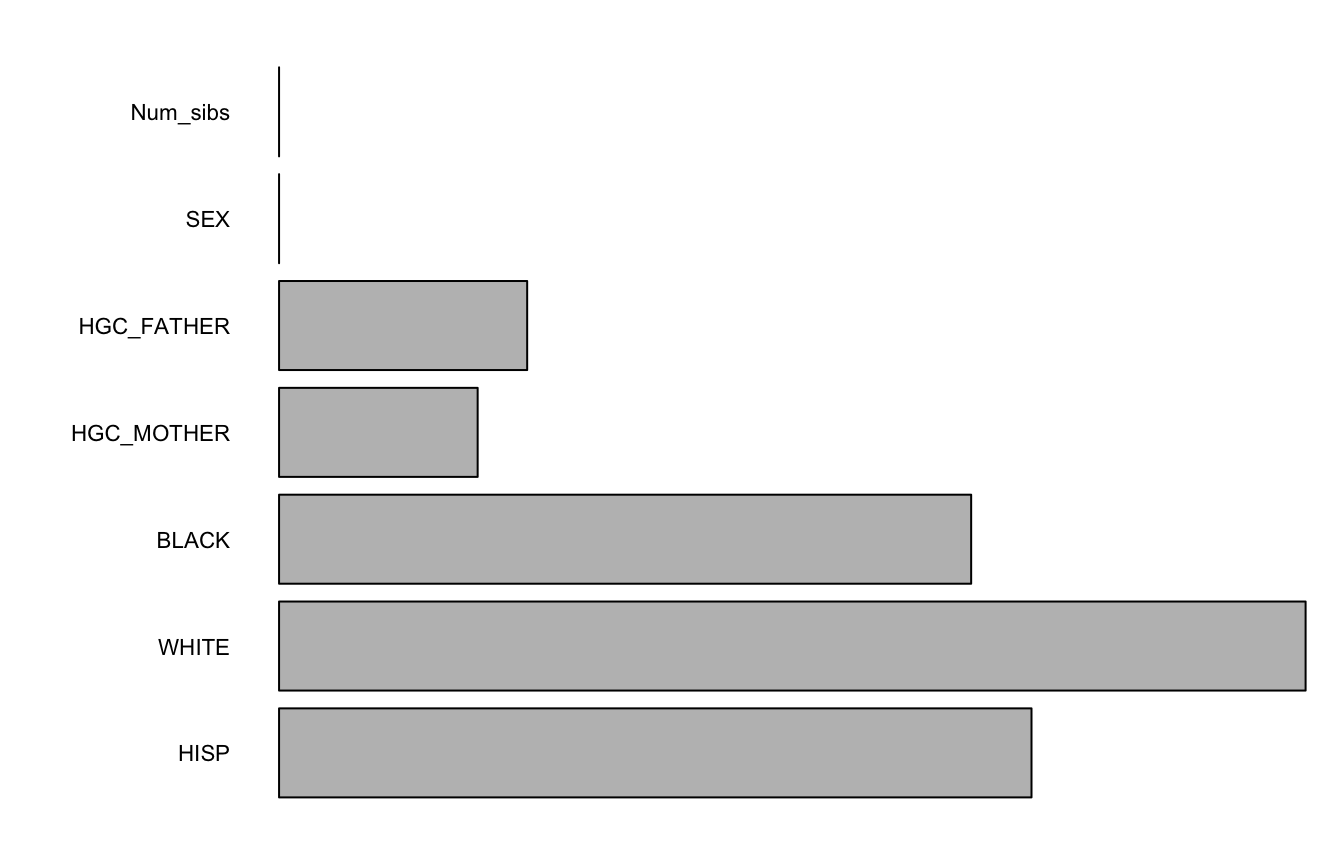
Example Forest
set.seed(1234)
forest1 <- splineForest(~HISP+WHITE+BLACK+HGC_MOTHER+HGC_FATHER+SEX+Num_sibs,
BMI ~ AGE, "ID", nlsySample, degree = 1, df=2, intercept = FALSE, ntree=50, prob=1/2)
varImps <- varImpCoeff(forest1)
plotImp(varImps[,3])