Description
Staged Event Trees.
Description
Creates and fits staged event tree probability models, which are probabilistic graphical models capable of representing asymmetric conditional independence statements for categorical variables. Includes functions to create, plot and fit staged event trees from data, as well as many efficient structure learning algorithms. References: Carli F, Leonelli M, Riccomagno E, Varando G (2022). <doi: 10.18637/jss.v102.i06>. Collazo R. A., Görgen C. and Smith J. Q. (2018, ISBN:9781498729604). Görgen C., Bigatti A., Riccomagno E. and Smith J. Q. (2018) <arXiv:1705.09457>. Thwaites P. A., Smith, J. Q. (2017) <arXiv:1510.00186>. Barclay L. M., Hutton J. L. and Smith J. Q. (2013) <doi:10.1016/j.ijar.2013.05.006>. Smith J. Q. and Anderson P. E. (2008) <doi:10.1016/j.artint.2007.05.004>.
README.md
stagedtrees
Citation
To cite stagedtrees in publications use:
Carli F, Leonelli M, Riccomagno E, Varando G (2022). “The R Package stagedtrees for Structural Learning of Stratified Staged Trees.” Journal of Statistical Software, 102(6), 1-30. doi: 10.18637/jss.v102.i06 (URL: https://doi.org/10.18637/jss.v102.i06).
@Article{,
title = {The {R} Package {stagedtrees} for Structural Learning of Stratified Staged Trees},
author = {Federico Carli and Manuele Leonelli and Eva Riccomagno and Gherardo Varando},
journal = {Journal of Statistical Software},
year = {2022},
volume = {102},
number = {6},
pages = {1--30},
doi = {10.18637/jss.v102.i06},
}
Overview
stagedtrees is a package that implements staged event trees, a class of probability models for categorical random variables.
Installation
# Install stable version from CRAN:
install.packages("stagedtrees")
# Or the development version from GitHub:
remotes::install_github("stagedtrees/stagedtrees")
Usage
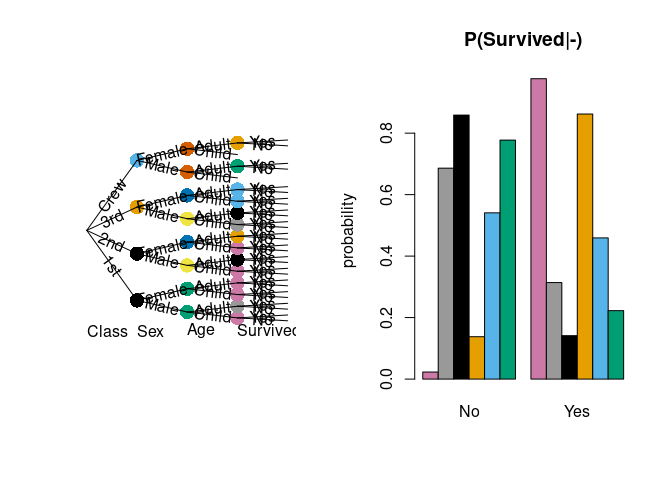
With the stagedtrees package it is possible to estimate (stratified) staged event trees from data, use them to compute probabilities, make predictions, visualize and compare different models.
library("stagedtrees")
tree <- Titanic |> full() |> stages_bhc() |> stndnaming(uniq = TRUE)
prob(tree, c(Survived="Yes"), conditional_on = c(Age="Adult"))
#> [1] 0.3107124
palette("Okabe-Ito")
par(mfrow = c(1,2))
plot(tree, col = "stages")
barplot(tree, var = "Survived", main = "P(Survived|-)", col = "stages")