Extract Statistics from Articles and Recompute P-Values.
statcheck 
What is statcheck?
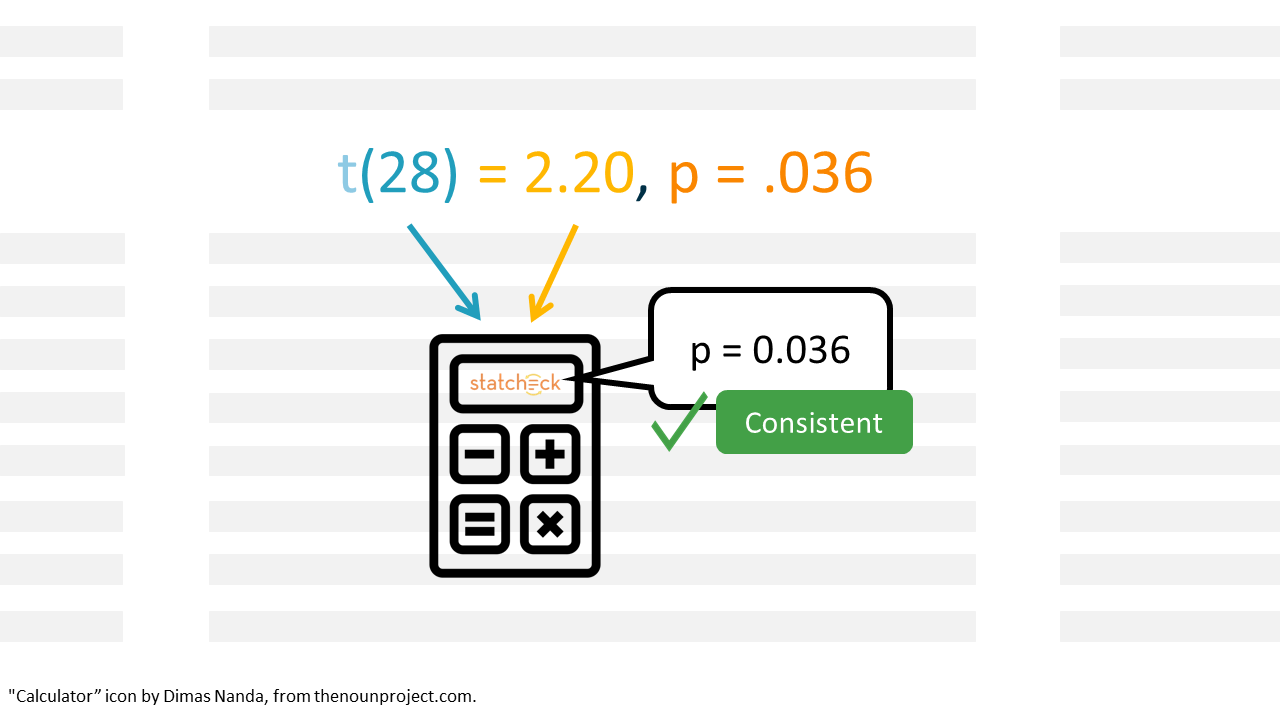
statcheck is a “spellchecker” for statistics. It checks whether your p-values match their accompanying test statistic and degrees of freedom.
statcheck searches for null-hypothesis significance test (NHST) in APA style (e.g., t(28) = 2.2, p < .05). It recalculates the p-value using the reported test statistic and degrees of freedom. If the reported and computed p-values don’t match, statcheck will flag the result as an error.
What can I use statcheck for?
statcheck is mainly useful for:
- Self-checks: you can use
statcheckto make sure your manuscript doesn’t contain copy-paste errors or other inconsistencies before you submit it to a journal. - Peer review: editors and reviewers can use
statcheckto check submitted manuscripts for statistical inconsistencies. They can ask authors for a correction or clarification before publishing a manuscript. - Research:
statcheckcan be used to automatically extract statistical test results from articles that can then be analyzed. You can for instance investigate whether you can predict statistical inconsistencies (see e.g., Nuijten et al., 2017 doi:10.1525/collabra.102), or use it to analyze p-value distributions (see e.g., Hartgerink et al., 2016 doi:10.7717/peerj.1935).
How does statcheck work?
The algorithm behind statcheck consists of four basic steps:
- Convert pdf and html articles to plain text files.
- Search the text for instances of NHST results. Specifically,
statcheckcan recognize t-tests, F-tests, correlations, z-tests, $\chi^2$ -tests, and Q-tests (from meta-analyses) if they are reported completely (test statistic, degrees of freedom, and p-value) and in APA style. - Recompute the p-value using the reported test statistic and degrees of freedom.
- Compare the reported and recomputed p-value. If the reported p-value does not match the computed one, the result is marked as an inconsistency (
errorin the output). If the reported p-value is significant and the computed is not, or vice versa, the result is marked as a gross inconsistency (decision_errorin the output).
statcheck takes into account correct rounding of the test statistic, and has the option to take into account one-tailed testing. See the manual for details.
Installation and use
For detailed information about installing and using statcheck, see the manual on RPubs.
Also see statcheck.io, a web-based interface for statcheck.
