Estimators and Plots for Gamma and Pareto Tail Detection.
Integration of C++ in Tailplots
Tailplots uses the R package Rcpp to integrate C++ code with R. One advantage of Rcpp is its extensive documentation, and many guides are available.
Structure of the Package
Under R/ you’ll find the package’s R code and two additional files, R/zzz.R and R/RcppExports.R. The first file sets the imports for the Tailplots package and can probably be removed now. The second contains the wrapper functions that connect the C++ code to R and shouldn’t be edited by hand (see the workflow below).
In the src/ folder you’ll find the C++ code. The only files that should be edited by hand are gamma.cpp and pareto.cpp; the other files are generated automatically by Rcpp. Any C++ function that you want to export and make available after devtools::load_all() must have the line // [[Rcpp::export]] immediately above the function definition. Include the following at the beginning of each C++ file:
#include <Rcpp.h>
using namespace Rcpp;
The rest is standard C++ code.
Note that the DESCRIPTION file contains:
Imports:
Rcpp
LinkingTo:
Rcpp
Workflow
The usual R package workflow described in R Packages (2e)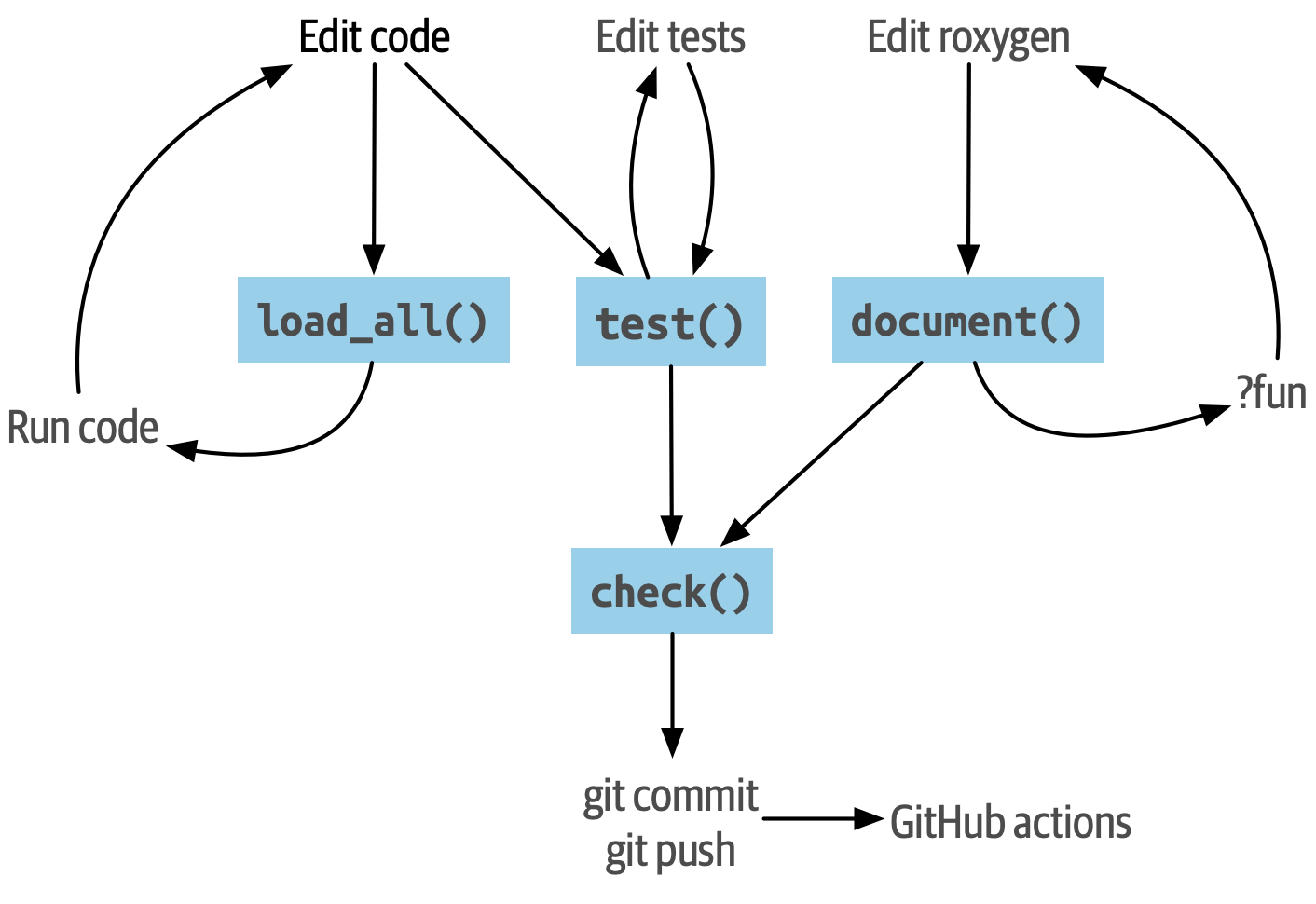 is supplemented by the command
is supplemented by the command Rcpp::compileAttributes(). To access newly written or modified C++ and R functions from the terminal, run:
Rcpp::compileAttributes()
devtools::load_all()
Everything else remains the same.