Fit and Tune Models to Detect Treatment Effect Heterogeneity.
tehtuner
The goal of tehtuner is to implement methods to fit models to detect and model treatment effect heterogeneity (TEH) while controlling the Type-I error of falsely detecting a differential effect when the conditional average treatment effect is uniform across the study population.
Currently tehtuner supports Virtual Twins models (Foster et al., 2011) for detecting TEH using the permutation procedure proposed in (Wolf et al., 2022).
Virtual Twins is a two-step approach to detecting differential treatment effects. Subjects’ conditional average treatment effects (CATEs) are first estimated in Step 1 using a flexible model. Then, a simple and interpretable model is fit in Step 2 to model these estimated CATEs as a function of the covariates.
The Step 2 model is dependent on some tuning parameter. This parameter is selected to control the Type-I error rate by permuting the data under the null hypothesis of a constant treatment effect and identifying the minimal null penalty parameter (MNPP), which is the smallest penalty parameter that yields a Step 2 model with no covariate effects. The $1-\alpha$ quantile of the distribution of is then used to fit the Step 2 model on the original data. In dong so, the Type-I error rate is controlled to be $\alpha$.
Installation
tehtuner is available on CRAN; you can download the release version with:
install.packages("tehtuner")
You can download the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("jackmwolf/tehtuner")
Example
We consider simulated data from a small clinical trial with 1000 subjects. Each subject has 10 measured covariates, 8 continuous and 2 binary. We are interested in estimating and understanding the CATE through Virtual Twins.
library(tehtuner)
data("tehtuner_example")
We will consider a Virtual Twins model using a random forest to estimate the CATEs in Step 1 and then fitting a regression tree on the estimated CATEs in Step 2 with the Type-I error rate set at $\alpha = 0.2$.
set.seed(100)
vt_cate <- tunevt(
data = tehtuner_example, Y = "Y", Trt = "Trt", step1 = "randomforest",
step2 = "rtree", alpha0 = 0.2, p_reps = 100, ntree = 50
)
The fitted Step 2 model can be accessed via $vtmod. In this case, as we used a regression tree in Step 2, our final model model is of class rpart.object.
vt_cate$vtmod
#> n= 1000
#>
#> node), split, n, deviance, yval
#> * denotes terminal node
#>
#> 1) root 1000 18372.4300 1.6830340
#> 2) V1< -3.008541 511 5643.7030 -0.3942647 *
#> 3) V1>=-3.008541 489 8219.4140 3.8537890
#> 6) V3>=0.000282894 19 448.7299 -5.3806930 *
#> 7) V3< 0.000282894 470 6084.9480 4.2270980 *
rpart.plot::rpart.plot(vt_cate$vtmod, digits = -2)
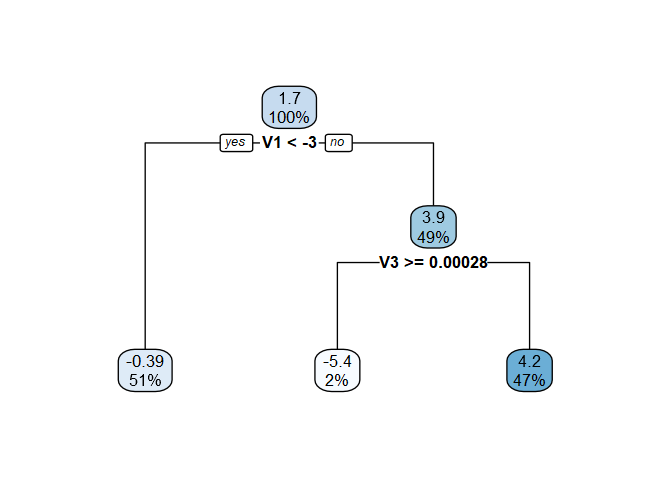
The fitted model for the CATE is a function of the covariates (V1, and V3), so we would conclude that there is treatment effect heterogeneity at the 20% level.
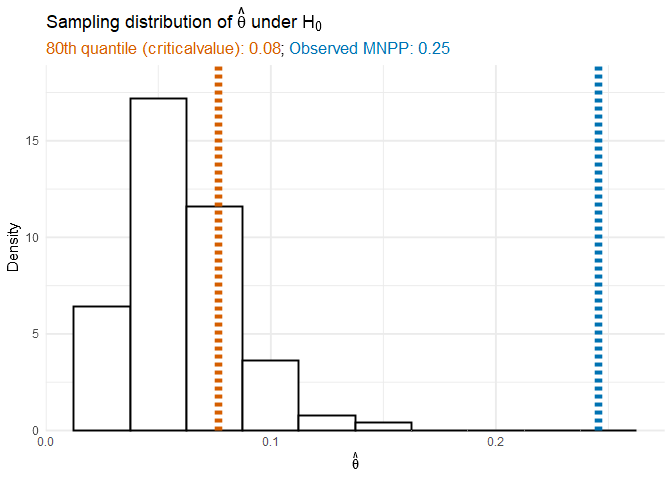
We can also look at the null distribution of the MNPP through vt_cate$theta_null. The 80th percentile of $\hat\theta$ under the null hypothesis is
quantile(vt_cate$theta_null, 0.8)
#> 80%
#> 0.07673669
while the MNPP of our observed data is
vt_cate$mnpp
#> [1] 0.2454389
The procedure fit the Step 2 model using the 80th quantile of the null distribution which resulted in a model that included covariates since the MNPP was above the 80th quantile.
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> Warning: The dot-dot notation (`..density..`) was deprecated in ggplot2 3.4.0.
#> ℹ Please use `after_stat(density)` instead.
Running in Parallel
Development version 0.1.1.9001 added the parallel option to tunevt() which allows the user to perform the permutation procedure in parallel to reduce computation times. Before doing so, you must register a parallel backend; see ?foreach::foreach for more information.
For example, to carry out 100 permutations across 2 processors:
cl <- parallel::makeCluster(2)
doParallel::registerDoParallel(cl)
vt_cate_parallel <- tunevt(
data = tehtuner_example, Y = "Y", Trt = "Trt", step1 = "randomforest",
step2 = "rtree", alpha0 = 0.2, p_reps = 100, ntree = 50, parallel = TRUE
)
parallel::stopCluster(cl)
References
Foster, J. C., Taylor, J. M., & Ruberg, S. J. (2011). Subgroup identification from randomized clinical trial data. Statistics in Medicine, 30(24), 2867–2880. https://doi.org/10.1002/sim.4322
Wolf, J. M., Koopmeiners, J. S., & Vock, D. M. (2022). A permutation procedure to detect heterogeneous treatment effects in randomized clinical trials while controlling the type-I error rate. Clinical Trials 19(5). https://doi.org/10.1177/17407745221095855
Deng C., Wolf J. M., Vock D. M., Carroll D. M., Hatsukami D. K., Leng N., & Koopmeiners J. S. (2023). “Practical guidance on modeling choices for the virtual twins method.” Journal of Biopharmaceutical Statistics. https://doi.org/10.1080/10543406.2023.2170404.