Competing Risks Estimation.
tidycmprsk 
The tidycmprsk package provides an intuitive interface for working with the competing risk endpoints. The package wraps the cmprsk package, and exports functions for univariate cumulative incidence estimates with cuminc() and competing risk regression with crr().
The package also includes broom-style tidiers: tidy(), augment(), and glance().
Installation
You can install {tidycmprsk} with the following code.
install.packages("tidycmprsk")
Install the development version of {tidycmprsk} with:
# install.packages("devtools")
devtools::install_github("MSKCC-Epi-Bio/tidycmprsk")
Competing Risk Regression
Fit a Fine and Gray competing risks regression model using the the example data, trial.
library(tidycmprsk)
crr_mod <- crr(Surv(ttdeath, death_cr) ~ age + trt, trial)
#> 11 cases omitted due to missing values
crr_mod
#>
#> ── crr() ───────────────────────────────────────────────────────────────────────
#> • Call Surv(ttdeath, death_cr) ~ age + trt
#> • Failure type of interest "death from cancer"
#>
#> Variable Coef SE HR 95% CI p-value
#> age 0.006 0.010 1.01 0.99, 1.03 0.56
#> trtDrug B 0.417 0.279 1.52 0.88, 2.62 0.13
The tidycmprsk plays well with other packages, such as gtsummary.
tbl <-
crr_mod %>%
gtsummary::tbl_regression(exponentiate = TRUE) %>%
gtsummary::add_global_p() %>%
add_n(location = "level")
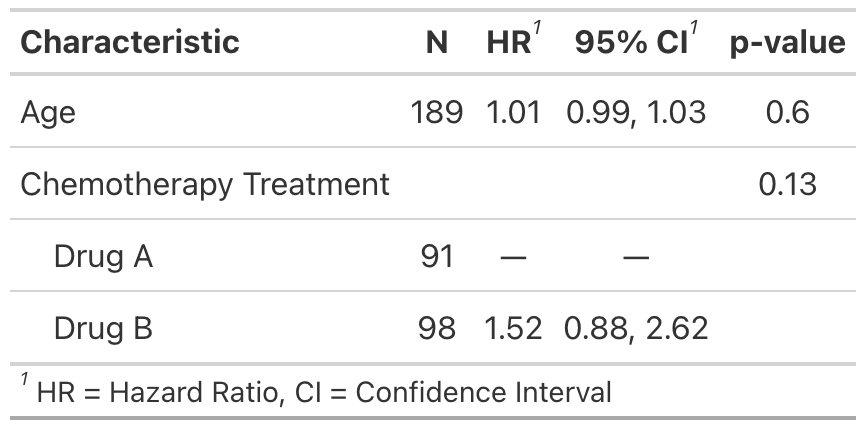
gtsummary::inline_text(tbl, variable = age)
#> [1] "1.01 (95% CI 0.99, 1.03; p=0.6)"
Cumulative Incidence
cuminc(Surv(ttdeath, death_cr) ~ 1, trial)
#>
#> ── cuminc() ────────────────────────────────────────────────────────────────────
#> • Failure type "death from cancer"
#> time n.risk estimate std.error 95% CI
#> 5.00 199 0.000 0.000 NA, NA
#> 10.0 189 0.030 0.012 0.012, 0.061
#> 15.0 158 0.120 0.023 0.079, 0.169
#> 20.0 116 0.215 0.029 0.161, 0.274
#> • Failure type "death other causes"
#> time n.risk estimate std.error 95% CI
#> 5.00 199 0.005 0.005 0.000, 0.026
#> 10.0 189 0.025 0.011 0.009, 0.054
#> 15.0 158 0.090 0.020 0.055, 0.135
#> 20.0 116 0.205 0.029 0.152, 0.264
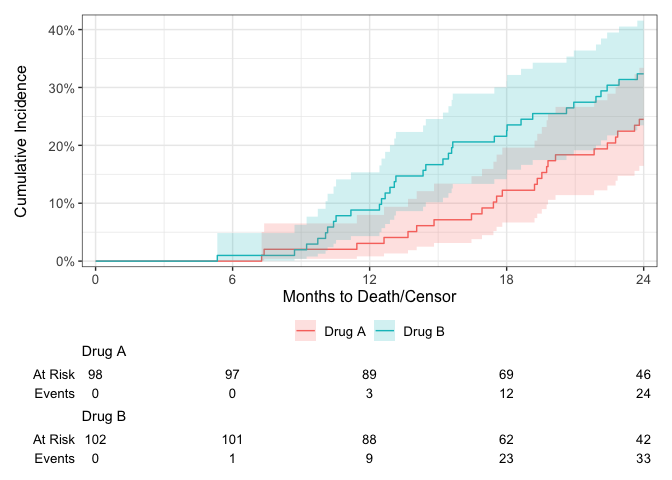
Plot risks using using the {ggsurvfit} package.
library(ggsurvfit)
#> Loading required package: ggplot2
cuminc(Surv(ttdeath, death_cr) ~ trt, trial) %>%
ggcuminc() +
add_confidence_interval() +
add_risktable() +
scale_ggsurvfit(x_scales = list(breaks = seq(0, 24, by = 6)))
#> Plotting outcome "death from cancer".
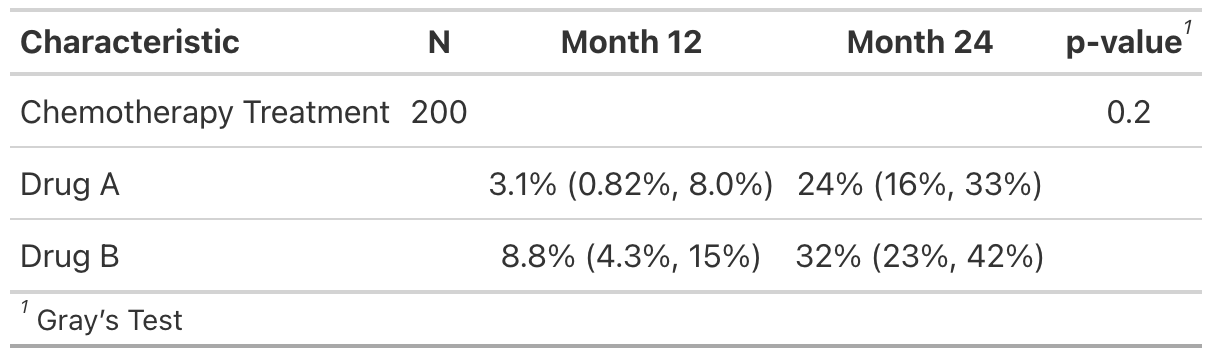
Summary table
tbl <-
cuminc(Surv(ttdeath, death_cr) ~ trt, trial) %>%
tbl_cuminc(times = c(12, 24), label_header = "**Month {time}**") %>%
add_p() %>%
add_n()
Contributing
Please note that the {tidycmprsk} project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms. Thank you to all contributors!
@abduazizR, @ABohynDOE, @ddsjoberg, @dublinQAQ, @erikvona, @fdehrich, @gustavomodelli, @hadley, @karissawhiting, @Lamenace23, @m-freitag, @mjtjmjtj, @nettam, @Nickrou10, @pteridin, @ramashka328, @SoumyaRo, @t-vinn, and @tengfei-emory
Limitations
The tidycmprsk package implements most features (and more) available in cmprsk. However, the time interaction features available in cmprsk::crr() is not available in tidycmprsk.