Add the 'dann' Model and the 'sub_dann' Model to the Tidymodels Ecosystem.
tidydann 
Package Introduction
In k nearest neighbors, the shape of the neighborhood is usually circular. Discriminant Adaptive Nearest Neighbors (dann) is a variation of k nearest neighbors where the shape of the neighborhood is data driven. The neighborhood is elongated along class boundaries and shrunk in the orthogonal direction. See Discriminate Adaptive Nearest Neighbor Classification by Hastie and Tibshirani.
This package brings the dann package into the tidymodels ecosystem.
Models:
- dann -> nearest_neighbor_adaptive with engine dann.
- sub_dann -> nearest_neighbor_adaptive with engine sub_dann.
Example 1: fit and predict with dann
In this example, simulated data is made. The overall trend is a circle inside a square.
knitr::opts_chunk$set(echo = TRUE, fig.width = 10, fig.height = 10)
library(parsnip)
library(rsample)
library(scales)
library(dials)
library(tune)
library(yardstick)
library(workflows)
library(tidydann)
library(dplyr, warn.conflicts = FALSE)
library(ggplot2)
library(mlbench)
# Create training data
set.seed(1)
circle_data <- mlbench.circle(700, 2) |>
tibble::as_tibble()
colnames(circle_data) <- c("X1", "X2", "Y")
set.seed(42)
split <- initial_split(circle_data, prop = .80)
train <- training(split)
test <- testing(split)
ggplot(train, aes(x = X1, y = X2, colour = as.factor(Y))) +
geom_point() +
labs(title = "Train Data", colour = "Y")
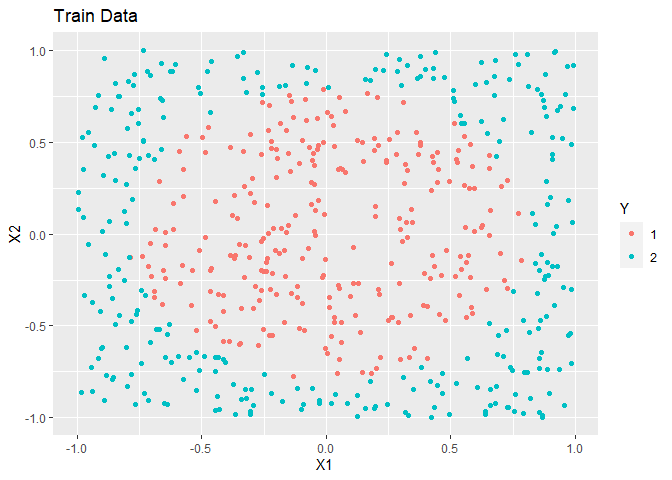
AUC is nearly perfect for these data.
model <- nearest_neighbor_adaptive(neighbors = 5, neighborhood = 50, matrix_diagonal = 1) |>
set_engine("dann") |>
fit(formula = Y ~ X1 + X2, data = train)
testPredictions <- model |>
predict(new_data = test, type = "prob")
testPredictions <- test |>
select(Y) |>
bind_cols(testPredictions)
testPredictions |>
roc_auc(truth = Y, event_level = "first", .pred_1)
#> # A tibble: 1 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 roc_auc binary 0.991
Example 2: cross validation with sub dann
In general, dann will struggle as unrelated variables are intermingled with informative variables. To deal with this, sub_dann projects the data onto a unique subspace and then calls dann on the subspace. In the below example there are 2 related variables and 5 that are unrelated.
######################
# Circle data with unrelated variables
######################
# Create training data
set.seed(1)
circle_data <- mlbench.circle(700, 2) |>
tibble::as_tibble()
colnames(circle_data) <- c("X1", "X2", "Y")
# Add 5 unrelated variables
circle_data <- circle_data |>
mutate(
U1 = runif(700, -1, 1),
U2 = runif(700, -1, 1),
U3 = runif(700, -1, 1),
U4 = runif(700, -1, 1),
U5 = runif(700, -1, 1)
)
set.seed(42)
split <- initial_split(circle_data, prop = .80)
train <- training(split)
test <- testing(split)
Without careful feature selection, dann’s performance suffers.
model <- nearest_neighbor_adaptive(neighbors = 5, neighborhood = 50, matrix_diagonal = 1) |>
set_engine("dann") |>
fit(formula = Y ~ ., data = train)
testPredictions <- model |>
predict(new_data = test, type = "prob")
testPredictions <- test |>
select(Y) |>
bind_cols(testPredictions)
testPredictions |>
roc_auc(truth = Y, event_level = "first", .pred_1)
#> # A tibble: 1 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 roc_auc binary 0.759
To deal with uninformative variables, a sub_dann model with tuned parameters is trained.
# define workflow
sub_dann_spec <-
nearest_neighbor_adaptive(
neighbors = tune(),
neighborhood = tune(),
matrix_diagonal = tune(),
weighted = tune(),
sphere = tune(),
num_comp = tune()
) |>
set_engine("sub_dann") |>
set_mode("classification")
sub_dann_wf <- workflow() |>
add_model(sub_dann_spec) |>
add_formula(Y ~ .)
# define grid
set.seed(2)
finalized_neighborhood <- neighborhood() |> get_n_frac(train, frac = .20)
finalized_num_comp <- num_comp() |> get_p(train[-1])
grid <- grid_random(
neighbors(),
finalized_neighborhood,
matrix_diagonal(),
weighted(),
sphere(),
finalized_num_comp,
size = 30,
filter = neighbors <= neighborhood
)
# tune
set.seed(123)
cv <- vfold_cv(data = train, v = 5)
sub_dann_tune_res <- sub_dann_wf |>
tune_grid(resamples = cv, grid = grid)
best_model <- sub_dann_tune_res |>
select_best(metric = "roc_auc")
With the best hyperparameters found, a final model on all training data is fit. Test AUC improved.
# retrain on all data
final_model <-
sub_dann_wf |>
finalize_workflow(best_model) |>
last_fit(split)
final_model |>
collect_metrics() |>
filter(.metric == "roc_auc") |>
select(.metric, .estimator, .estimate)
#> # A tibble: 1 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 roc_auc binary 0.985