Run Predictions Inside the Database.
tidypredict 
The main goal of tidypredict is to enable running predictions inside databases. It reads the model, extracts the components needed to calculate the prediction, and then creates an R formula that can be translated into SQL. In other words, it is able to parse a model such as this one:
model <- lm(mpg ~ wt + cyl, data = mtcars)
tidypredict can return a SQL statement that is ready to run inside the database. Because it uses dplyr’s database interface, it works with several databases back-ends, such as MS SQL:
tidypredict_sql(model, dbplyr::simulate_mssql())
## <SQL> (39.686261480253 + (`wt` * -3.19097213898374)) + (`cyl` * -1.5077949682598)
Installation
Install tidypredict from CRAN using:
install.packages("tidypredict")
Or install the development version using devtools as follows:
install.packages("remotes")
remotes::install_github("tidymodels/tidypredict")
Functions
tidypredict has only a few functions, and it is not expected that number to grow much. The main focus at this time is to add more models to support.
| Function | Description |
|---|---|
tidypredict_fit() | Returns an R formula that calculates the prediction |
tidypredict_sql() | Returns a SQL query based on the formula from tidypredict_fit() |
tidypredict_to_column() | Adds a new column using the formula from tidypredict_fit() |
tidypredict_test() | Tests tidyverse predictions against the model’s native predict() function |
tidypredict_interval() | Same as tidypredict_fit() but for intervals (only works with lm and glm) |
tidypredict_sql_interval() | Same as tidypredict_sql() but for intervals (only works with lm and glm) |
parse_model() | Creates a list spec based on the R model |
as_parsed_model() | Prepares an object to be recognized as a parsed model |
How it works
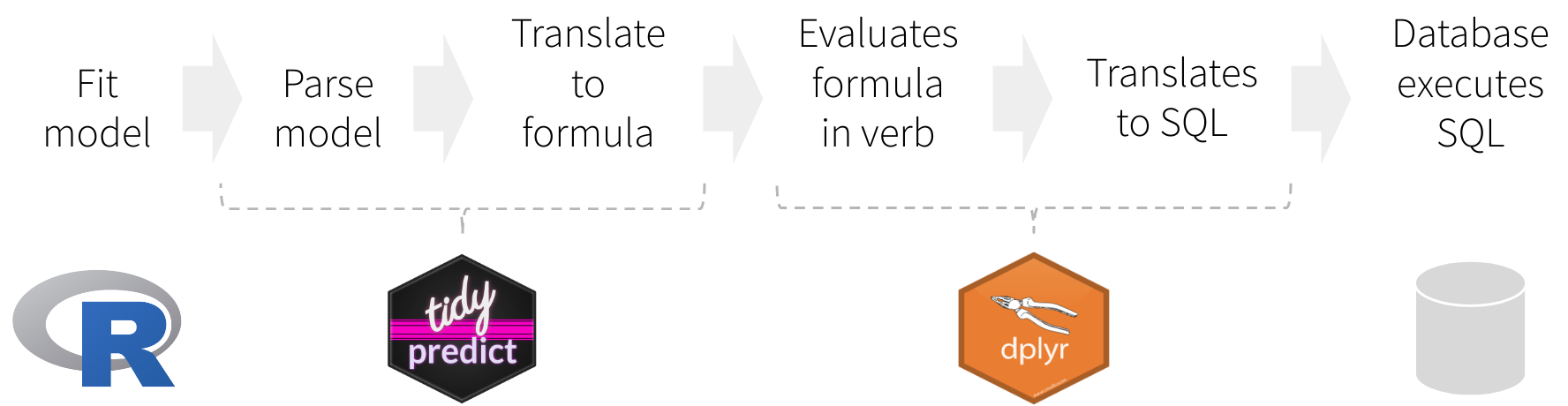
Instead of translating directly to a SQL statement, tidypredict creates an R formula. That formula can then be used inside dplyr. The overall workflow would be as illustrated in the image above, and described here:
- Fit the model using a base R model, or one from the packages listed in Supported Models
tidypredictreads model, and creates a list object with the necessary components to run predictionstidypredictbuilds an R formula based on the list objectdplyrevaluates the formula created bytidypredictdplyrtranslates the formula into a SQL statement, or any other interfaces.- The database executes the SQL statement(s) created by
dplyr
Parsed model spec
tidypredict writes and reads a spec based on a model. Instead of simply writing the R formula directly, splitting the spec from the formula adds the following capabilities:
- No more saving models as
.rds- Specifically for cases when the model needs to be used for predictions in a Shiny app. - Beyond R models - Technically, anything that can write a proper spec, can be read into
tidypredict. It also means, that the parsed model spec can become a good alternative to using PMML.
Supported models
The following models are supported by tidypredict:
- Linear Regression -
lm() - Generalized Linear model -
glm() - Random Forest models -
randomForest::randomForest() - Random Forest models, via
ranger-ranger::ranger() - MARS models -
earth::earth() - XGBoost models -
xgboost::xgb.Booster.complete() - Cubist models -
Cubist::cubist() - Tree models, via
partykit-partykit::ctree()
parsnip
tidypredict supports models fitted via the parsnip interface. The ones confirmed currently work in tidypredict are:
lm()-parsnip:linear_reg()with “lm” as the engine.randomForest::randomForest()-parsnip:rand_forest()with “randomForest” as the engine.ranger::ranger()-parsnip:rand_forest()with “ranger” as the engine.earth::earth()-parsnip:mars()with “earth” as the engine.
broom
The tidy() function from broom works with linear models parsed via tidypredict
pm <- parse_model(lm(wt ~ ., mtcars))
tidy(pm)
## # A tibble: 11 × 2
## term estimate
## <chr> <dbl>
## 1 (Intercept) -0.231
## 2 mpg -0.0417
## 3 cyl -0.0573
## 4 disp 0.00669
## 5 hp -0.00323
## 6 drat -0.0901
## 7 qsec 0.200
## 8 vs -0.0664
## 9 am 0.0184
## 10 gear -0.0935
## 11 carb 0.249
Contributing
This project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
For questions and discussions about tidymodels packages, modeling, and machine learning, please post on Posit Community.
If you think you have encountered a bug, please submit an issue.
Either way, learn how to create and share a reprex (a minimal, reproducible example), to clearly communicate about your code.
Check out further details on contributing guidelines for tidymodels packages and how to get help.