Description
Utilities to Retrieve Rulelists from Model Fits, Filter, Prune, Reorder and Predict on Unseen Data.
Description
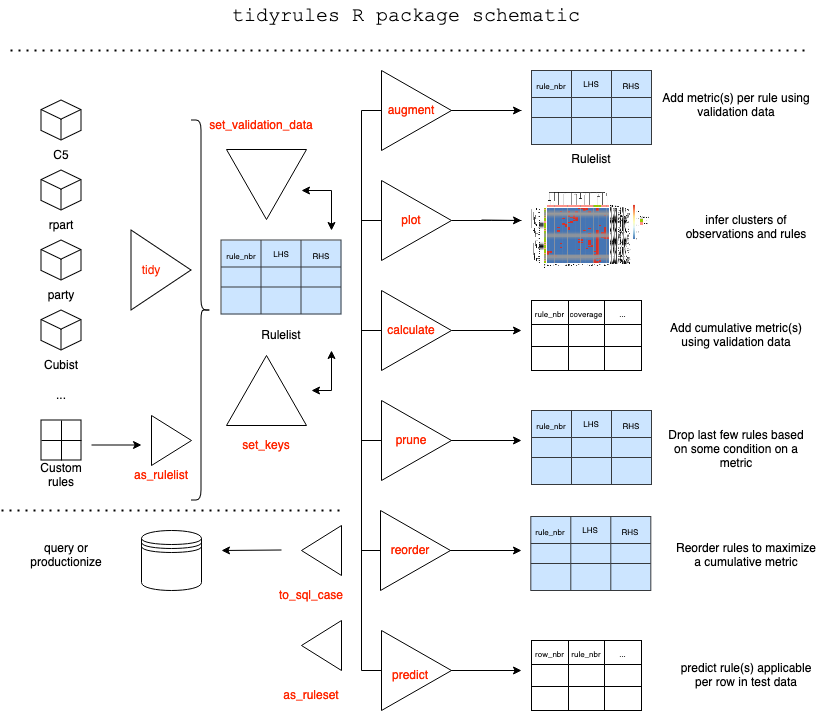
Provides a framework to work with decision rules. Rules can be extracted from supported models, augmented with (custom) metrics using validation data, manipulated using standard dataframe operations, reordered and pruned based on a metric, predict on unseen (test) data. Utilities include; Creating a rulelist manually, Exporting a rulelist as a SQL case statement and so on. The package offers two classes; rulelist and ruleset based on dataframe.
README.md
tidyrules
tidyrulesRpackage provides a framework to work with decision rules. Rules can be extracted from supported models, augmented with (custom) metrics using validation data, manipulated using standard dataframe operations, reordered and pruned based on a metric, predict on unseen (test) data. Utilities include; Creating a rulelist manually, Exporting a rulelist as a SQL case statement and so on. The package offers two classes; rulelist and ruleset based on dataframe.
website: https://talegari.github.io/tidyrules/
Example
expand/collapse
library(tidyrules)
model_c5 = C50::C5.0(Species ~ ., data = iris, rules = TRUE)
pander::pandoc.table(tidy(model_c5), split.tables = 120)
#>
#> ----------------------------------------------------------------------------------------------
#> rule_nbr trial_nbr LHS RHS support confidence lift
#> ---------- ----------- ---------------------------- ------------ --------- ------------ ------
#> 1 1 ( Petal.Length <= 1.9 ) setosa 50 0.9808 2.9
#>
#> 2 1 ( Petal.Length > 1.9 ) & ( versicolor 48 0.96 2.9
#> Petal.Length <= 4.9 ) & (
#> Petal.Width <= 1.7 )
#>
#> 3 1 ( Petal.Width > 1.7 ) virginica 46 0.9583 2.9
#>
#> 4 1 ( Petal.Length > 4.9 ) virginica 46 0.9375 2.8
#> ----------------------------------------------------------------------------------------------